
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
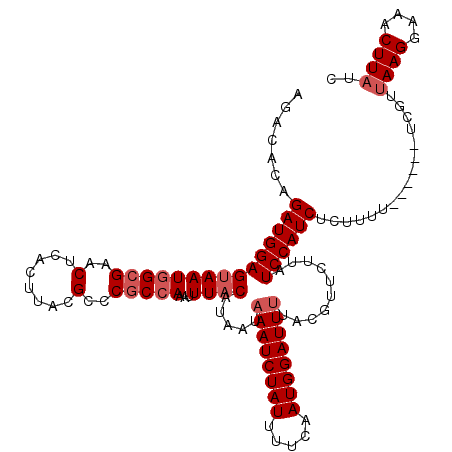
| Location | 3,395,403 – 3,395,517 |
| Length | 114 |
| Max. P | 0.862018 |

| Location | 3,395,403 – 3,395,517 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -19.45 |
| Energy contribution | -20.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
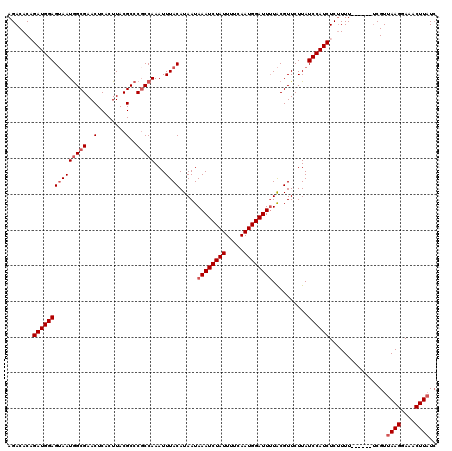
>3R_DroMel_CAF1 3395403 114 + 27905053 AGACACAGAUGGAGUAAUGGCGAACUCACUUACGCCCGCCAAAUUUACAUAAUAAAUCUAUUUUCAAUGGAUUCUACGUUCUUAUCCAUCUCUUUU------UCGUUAAGGAAACUUAUC ......((((((((((((((((..(........)..)))))...))))......(((((((.....)))))))...........))))))).....------....((((....)))).. ( -25.10) >DroSim_CAF1 4300 114 + 1 AGACACAGAUGGAGUAAUGGCGAACUCACUUACGCCCGCCAAAUUUACAUAAUAAAUCUAUUUUCAAUGGAUUUUACGUUCUUAUCCAUCUCUUUU------UCGUUAAGGAAACUUAUC ......((((((((((((((((..(........)..)))))...)))).....((((((((.....))))))))..........))))))).....------....((((....)))).. ( -25.90) >DroEre_CAF1 2680 120 + 1 AGACACUGAUGGAGAAAUGGCGAACUCACUUACGCCCGCCAAAUUUACAUAAUAAAUCUAUUUUCAAUGGAUUUUGCGUUCUUAUCCAUCUCGUUUUCGCUUUCGUUAAGGAAACUUAUC ((((...((((((....(((((..(........)..)))))............((((((((.....))))))))..........))))))..))))..........((((....)))).. ( -24.50) >DroYak_CAF1 8083 114 + 1 AGACACUGAUGGAGUAAUAGCGAACUCACUUACGCCCUCCAAAUUUACAUAAUAAAUCUAUUUGCAAUGGAUUUUACGUUCUUAUCCAUCUCUUUU------CCGUUAAGGAAACUUUUA .......((((((.(((.((((....................(((.....)))((((((((.....))))))))..)))).)))))))))...(((------((.....)))))...... ( -18.80) >consensus AGACACAGAUGGAGUAAUGGCGAACUCACUUACGCCCGCCAAAUUUACAUAAUAAAUCUAUUUUCAAUGGAUUUUACGUUCUUAUCCAUCUCUUUU______UCGUUAAGGAAACUUAUC .......(((((((((((((((..(........)..)))))...)))).....((((((((.....))))))))..........))))))................((((....)))).. (-19.45 = -20.70 + 1.25)

| Location | 3,395,403 – 3,395,517 |
|---|---|
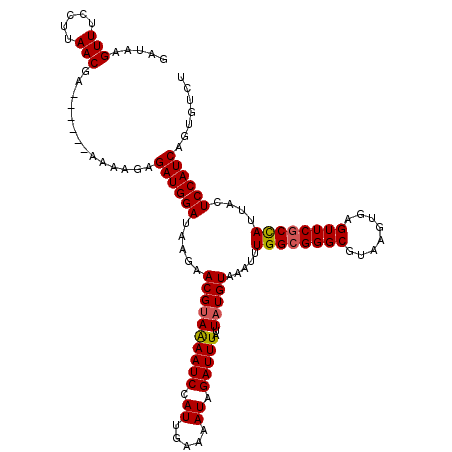
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
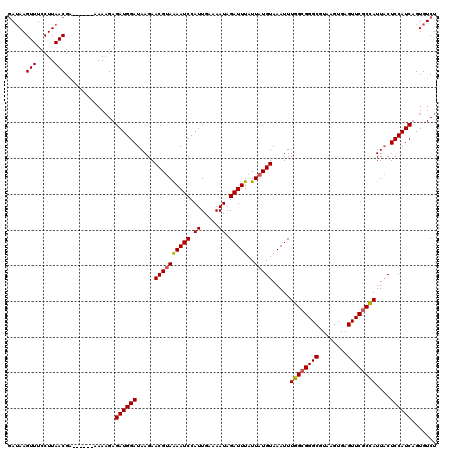
>3R_DroMel_CAF1 3395403 114 - 27905053 GAUAAGUUUCCUUAACGA------AAAAGAGAUGGAUAAGAACGUAGAAUCCAUUGAAAAUAGAUUUAUUAUGUAAAUUUGGCGGGCGUAAGUGAGUUCGCCAUUACUCCAUCUGUGUCU ..((((....))))..((------...(.(((((((.....((((((((((...(....)..)))))..))))).....((((((((........))))))))....))))))).).)). ( -28.00) >DroSim_CAF1 4300 114 - 1 GAUAAGUUUCCUUAACGA------AAAAGAGAUGGAUAAGAACGUAAAAUCCAUUGAAAAUAGAUUUAUUAUGUAAAUUUGGCGGGCGUAAGUGAGUUCGCCAUUACUCCAUCUGUGUCU ..((((....))))..((------...(.(((((((.....((((((((((...(....)..))))..)))))).....((((((((........))))))))....))))))).).)). ( -27.60) >DroEre_CAF1 2680 120 - 1 GAUAAGUUUCCUUAACGAAAGCGAAAACGAGAUGGAUAAGAACGCAAAAUCCAUUGAAAAUAGAUUUAUUAUGUAAAUUUGGCGGGCGUAAGUGAGUUCGCCAUUUCUCCAUCAGUGUCU ((((..(((((((.....))).))))....((((((.(((.....................((((((((...))))))))(((((((........))))))).))).))))))..)))). ( -26.60) >DroYak_CAF1 8083 114 - 1 UAAAAGUUUCCUUAACGG------AAAAGAGAUGGAUAAGAACGUAAAAUCCAUUGCAAAUAGAUUUAUUAUGUAAAUUUGGAGGGCGUAAGUGAGUUCGCUAUUACUCCAUCAGUGUCU ......(((((.....))------)))...((((((.....((((....(((((((((((((....)))).)))))...))))..)))).((((....)))).....))))))....... ( -25.70) >consensus GAUAAGUUUCCUUAACGA______AAAAGAGAUGGAUAAGAACGUAAAAUCCAUUGAAAAUAGAUUUAUUAUGUAAAUUUGGCGGGCGUAAGUGAGUUCGCCAUUACUCCAUCAGUGUCU .....(((.....)))..............((((((.....((((((((((.((.....)).)))))..))))).....((((((((........))))))))....))))))....... (-21.62 = -21.75 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:52 2006