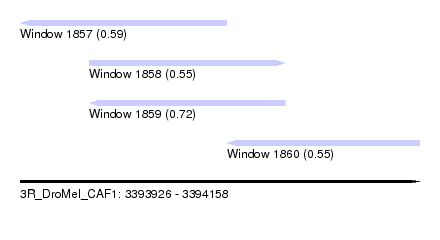
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,393,926 – 3,394,158 |
| Length | 232 |
| Max. P | 0.721085 |

| Location | 3,393,926 – 3,394,046 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3393926 120 - 27905053 GGUGAGCAAAAAAUGCUAUACGCAUUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGCCGGCAAUUAGUUUGUAUUAUUUUUCAGUUUACUUUGCCAUUUAUAUUUGUUCUUUUUGCC ((((((((((...(((((((.....))))))).......((..((((((((...........((((.....))))..........))))))))..))........))))))).....))) ( -24.50) >DroSim_CAF1 2848 120 - 1 GCUGAGCAAAAAAUGCUAUACACAUUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGCCGGCAAUUAGUUUGUAUUAUUUUUCAGUUUACUUUGCCAUUUAUAUUUGUUCUUUUAGCC ((((((((((...(((((((.....))))))).......((..((((((((...........((((.....))))..........))))))))..))........)))))))....))). ( -25.40) >DroEre_CAF1 1006 120 - 1 GCUGAGCAAAAAAUGUUUUAUACACUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGAUGGCAAUUAGUUUGUAUUAUUUUUCAGUUUACCUUGCCAUUUAUAUUUGUUCUUGUUGUC ...(((((((...((((.((.....)).)))).......((.(((((((((.......((((.(((.....))))))).......))))))))).))........)))))))........ ( -21.24) >DroYak_CAF1 6511 120 - 1 GCUGAGCAAAAAAUGUUAUAUACACUAUAACAGUUUUUCGUUGGGUAAACUUUAUUUGGACGGCAAUUAGUUUGUAUUAUUUUUCAGUUUACUUUGCCAUUUAUAUUUGUUCUUUUUGCA ((.(((((((...(((((((.....))))))).......((..((((((((...........((((.....))))..........))))))))..))........))))))).....)). ( -23.40) >consensus GCUGAGCAAAAAAUGCUAUACACACUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGACGGCAAUUAGUUUGUAUUAUUUUUCAGUUUACUUUGCCAUUUAUAUUUGUUCUUUUUGCC ...(((((((...(((((((.....))))))).......((..((((((((...........((((.....))))..........))))))))..))........)))))))........ (-23.48 = -23.10 + -0.38)

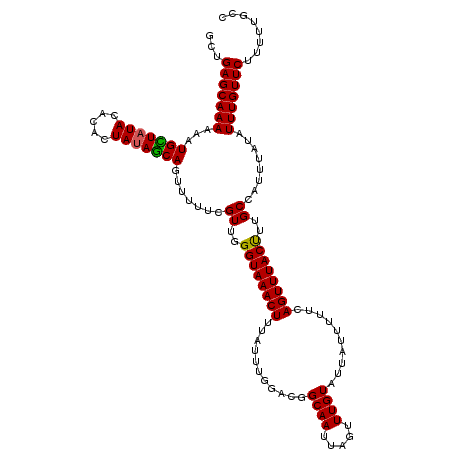
| Location | 3,393,966 – 3,394,080 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -16.77 |
| Consensus MFE | -13.05 |
| Energy contribution | -12.18 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3393966 114 + 27905053 AUAAUACAAACUAAUUGCCGGCCAAAUAAAGUUUACCCAACGAAAAACUGCUAUAAUGCGUAUAGCAUUUUUUGCUCACCCACUUUUCUUCGAUUUCGUUUAAACUUUUCCAAC------ ...................((......((((((((...(((((((....((......))....((((.....)))).................))))))))))))))).))...------ ( -15.90) >DroSim_CAF1 2888 120 + 1 AUAAUACAAACUAAUUGCCGGCCAAAUAAAGUUUACCCAACGAAAAACUGCUAUAAUGUGUAUAGCAUUUUUUGCUCAGCCACUUUUCUUCGAUUUCGUUUAAACUUUUCCAACGGCAAA ..............((((((.......((((((((...(((((((...(((((((.....)))))))......((...)).............))))))))))))))).....)))))). ( -25.50) >DroEre_CAF1 1046 114 + 1 AUAAUACAAACUAAUUGCCAUCCAAAUAAAGUUUACCCAACGAAAAACUGCUAUAGUGUAUAAAACAUUUUUUGCUCAGCCACUUUUCUUCGAUUUCGUUUAAACUUUUCCAAC------ ...........................((((((((...(((((((....((...(((((.....)))))....)).......(........).)))))))))))))))......------ ( -11.70) >DroYak_CAF1 6551 114 + 1 AUAAUACAAACUAAUUGCCGUCCAAAUAAAGUUUACCCAACGAAAAACUGUUAUAGUGUAUAUAACAUUUUUUGCUCAGCUACUUUUCUUCGAUUUCGUUUAAACUUUUCCAAC------ ...........................((((((((...(((((((...(((((((.....)))))))......((...)).............)))))))))))))))......------ ( -14.00) >consensus AUAAUACAAACUAAUUGCCGGCCAAAUAAAGUUUACCCAACGAAAAACUGCUAUAAUGUAUAUAACAUUUUUUGCUCAGCCACUUUUCUUCGAUUUCGUUUAAACUUUUCCAAC______ ...........................((((((((...(((((((....((...(((((.....)))))....)).......(........).)))))))))))))))............ (-13.05 = -12.18 + -0.88)

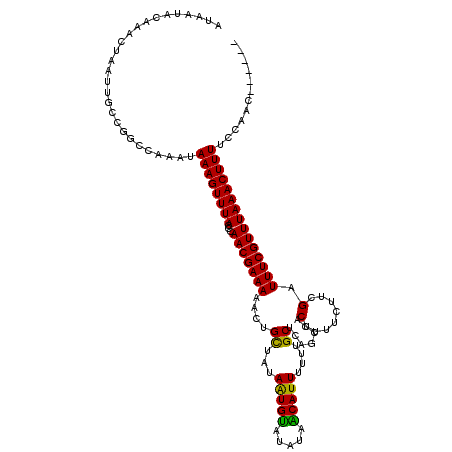
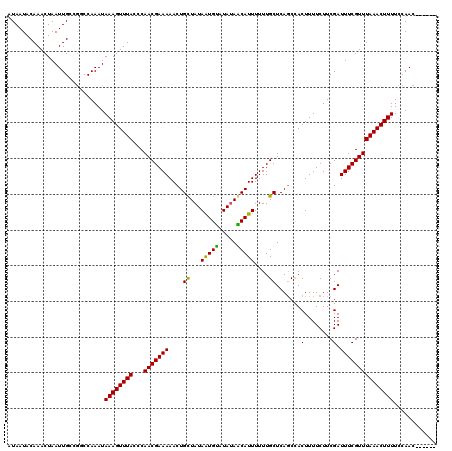
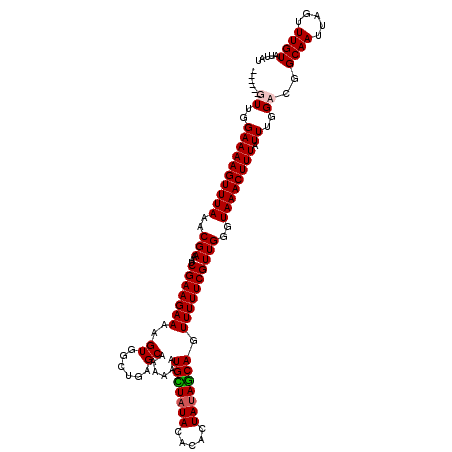
| Location | 3,393,966 – 3,394,080 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3393966 114 - 27905053 ------GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUGGGUGAGCAAAAAAUGCUAUACGCAUUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGCCGGCAAUUAGUUUGUAUUAU ------((..((((((((((..(((...(((((((.((((.(((((((.....)))).))).)))).......))))))))))..)))))))).))..))..((((.....))))..... ( -27.20) >DroSim_CAF1 2888 120 - 1 UUUGCCGUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUGGCUGAGCAAAAAAUGCUAUACACAUUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGCCGGCAAUUAGUUUGUAUUAU .(((((((..((((((((((..(((...(((((((..((......))......(((((((.....))))))).))))))))))..)))))))).))..).)))))).............. ( -32.40) >DroEre_CAF1 1046 114 - 1 ------GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUGGCUGAGCAAAAAAUGUUUUAUACACUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGAUGGCAAUUAGUUUGUAUUAU ------.(..((((((((((..(((...(((((((.((((...(((((.....)))))....)))).......))))))))))..)))))))).))..)...((((.....))))..... ( -21.60) >DroYak_CAF1 6551 114 - 1 ------GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUAGCUGAGCAAAAAAUGUUAUAUACACUAUAACAGUUUUUCGUUGGGUAAACUUUAUUUGGACGGCAAUUAGUUUGUAUUAU ------(((.((((((((((..(((...(((((((.....((...))......(((((((.....))))))).))))))))))..)))))))..))).))).((((.....))))..... ( -22.20) >consensus ______GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUGGCUGAGCAAAAAAUGCUAUACACACUAUAGCAGUUUUUCGUUGGGUAAACUUUAUUUGGACGGCAAUUAGUUUGUAUUAU ......((..((((((((((..(((...(((((((..((......))......(((((((.....))))))).))))))))))..)))))))).))..))..((((.....))))..... (-22.24 = -22.55 + 0.31)

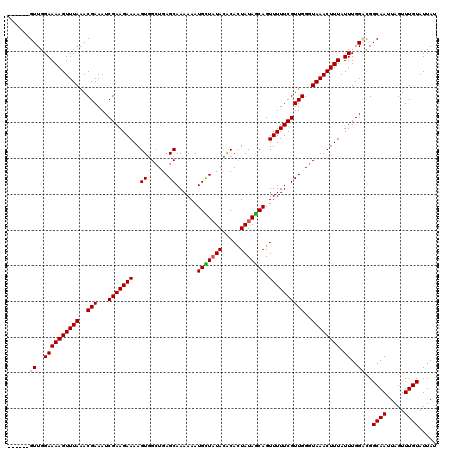
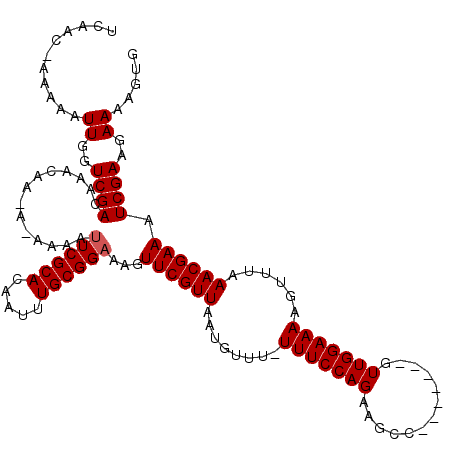
| Location | 3,394,046 – 3,394,158 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.09 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3394046 112 - 27905053 UGAACUAAAAAUUGGUCGACAUACAAAAAAAAAAUCGCACAAUUUGCGGAAAGUUCGUUAAUGUUU-UUUCCAGAAGCC-------GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUG ...........((..((((...............(((((.....)))))....((((((.....((-(((((((.....-------.)))))))))....)))))).))))..))..... ( -20.40) >DroSim_CAF1 2968 118 - 1 UCAACUAAAAAUUGGUCGACAUACAAAA--AAAUUCGCACAAUUUGCGGAAAGUUCGUUAAUGUUUUUUUCCAGAAGCCGUUUGCCGUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUG ...........((..((((.........--...((((((.....))))))...((((((..((..(((((((((..((.....))..)))))))))..)))))))).))))..))..... ( -24.90) >DroEre_CAF1 1126 108 - 1 UCAAC-AGAAAUUGGUCGACGAACAA---AAAAUUCGCACAAUUUGCGGAAAGUUCGUUAAUGUUU-UUUCCAGAAGCC-------GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUG .....-....(((..((..(((....---....((((((.....))))))...((((((.....((-(((((((.....-------.)))))))))....)))))).)))..))..))). ( -23.60) >DroYak_CAF1 6631 110 - 1 UUAAC-AAAAAUUGGUCGACGACCAA-AAAAAAUUCGCACAAUUUGCGGAAAGUUCGUUAAUGUUU-UUUCCAGAAGCC-------GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUA .....-.....((((((...))))))-......((((((.....))))))...((((((.....((-(((((((.....-------.)))))))))....)))))).............. ( -24.00) >consensus UCAAC_AAAAAUUGGUCGACAAACAA_A_AAAAUUCGCACAAUUUGCGGAAAGUUCGUUAAUGUUU_UUUCCAGAAGCC_______GUUGGAAAAGUUUAAACGAAAUCGAAGAAAAGUG ...........((..((((..............((((((.....))))))...((((((........(((((((.............)))))))......)))))).))))..))..... (-16.83 = -17.09 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:50 2006