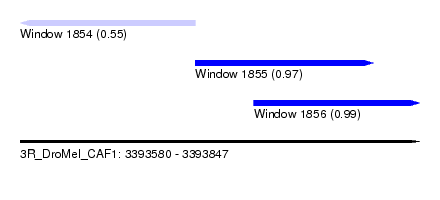
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,393,580 – 3,393,847 |
| Length | 267 |
| Max. P | 0.991340 |

| Location | 3,393,580 – 3,393,697 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -22.11 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3393580 117 - 27905053 CAAAGUUUGUCCAUUGUCUUUUUCUCAUUGUUGAGUAAAAAGUUUGCACUCUUAUUCAGCAGCCAGAUAAACUCAAAUCCAAUUGGCAACAGCUAUGAAUAACAUUUCAUGUAGACC--- ........(((..(((..(((.(((..(((((((((((..(((....))).)))))))))))..))).)))..)))......(((....))).((((((......))))))..))).--- ( -21.70) >DroSim_CAF1 2496 117 - 1 CAAAGUUUGUCCAUUGUCUUUUUCUCAUUGUUGAGUAAAAAGUUUGCACUCUCAUUCAGCAGCCAGAUAAACUCAAAUCCAAUUGGCAACAGCUAUGAAUAACAUUUCAUGUAGACC--- ...((((((((......((((((((((....)))).)))))).((((...........))))...)))))))).........(((....))).((((((......))))))......--- ( -21.20) >DroEre_CAF1 653 120 - 1 CAAAGUUUGCCCAUUGUCUUUUUGUCGUUGCUGAGUAAAAACUUUGCACUCUCAUUCAGCAGCCAGAUAAACUCAAAUCCAAUUGGCAACAGCUAUGAAUAACAUUUCAUUUAGACCUUG ...((((((((.((((..((((((((((((((((((.................))))))))))..)))))....)))..)))).)))))..)))(((((......))))).......... ( -25.93) >DroYak_CAF1 6178 103 - 1 CAAAGUUUGCCCAUUGUCUUUUUGCCAUUGUUGAGUGAAAACUUUGCACUCUCAUUCAGUAGCCAGAUAAACUCUAAUCCAAUUGGCAACAGCUAUGAAUAAC----------------- ((.((((((((.((((.......((.((((..(((((.........))))).....)))).)).(((.....)))....)))).)))))..))).))......----------------- ( -19.60) >consensus CAAAGUUUGCCCAUUGUCUUUUUCUCAUUGUUGAGUAAAAACUUUGCACUCUCAUUCAGCAGCCAGAUAAACUCAAAUCCAAUUGGCAACAGCUAUGAAUAACAUUUCAUGUAGACC___ ((.((((((((.((((..((((((((.(((((((((((.............)))))))))))...))))))....))..)))).)))))..))).))....................... (-17.25 = -17.75 + 0.50)

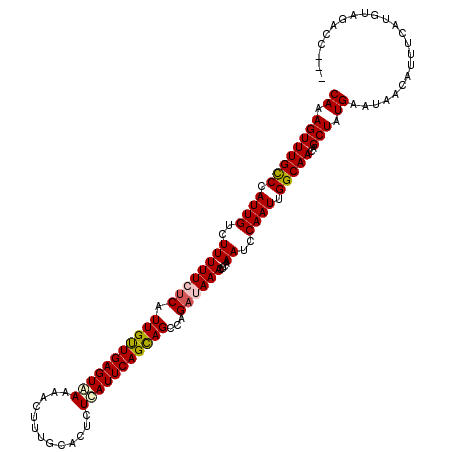
| Location | 3,393,697 – 3,393,816 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3393697 119 + 27905053 UUAUUUCUGGUU-UUUCUGCCACGGGAAUUGUAACGAAACCAUACCAAACGCGGAAAGCAAAAAGAAAGAUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUA .......(((((-((..(((..........)))..)))))))......((((.(((.((....((((((.(((((((......)))))))...))))))...)).))).....))))... ( -25.60) >DroSim_CAF1 2613 119 + 1 UUAUUUCUGGUU-UUUCUGCCACGGGAAUUGUAACGAAACCAAACCAAACGCGGAAAGCAAAAAGAAAGAUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUA .......(((((-((..(((..........)))..)))))))......((((.(((.((....((((((.(((((((......)))))))...))))))...)).))).....))))... ( -25.80) >DroEre_CAF1 773 114 + 1 UUAUUUCUGGUU-UUUGUGCCACGGGAAUUGUAAGGAAAC-----CAAACGCGGGAAGCAAAAAGAAAGGUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUA .......(((((-((..(((..........)))..)))))-----)).((((..(((((....((((((.(((((((......)))))))...))))))...)).))).....))))... ( -29.80) >DroYak_CAF1 6281 115 + 1 UUAUUUCUGGUUUUUUGUGCCACGGGAAUAGUAACGAAAC-----CAAACGCGGGAAGCAAAAAGAAAGGUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUA .........((((((((((((((((..............)-----)....((.....))....((((((.(((((((......)))))))...))))))))))))..))))))))..... ( -30.94) >consensus UUAUUUCUGGUU_UUUCUGCCACGGGAAUUGUAACGAAAC_____CAAACGCGGAAAGCAAAAAGAAAGAUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUA ...(..(((((.......)))).)..).....................((((.(((.((....((((((.(((((((......)))))))...))))))...)).))).....))))... (-25.80 = -25.55 + -0.25)

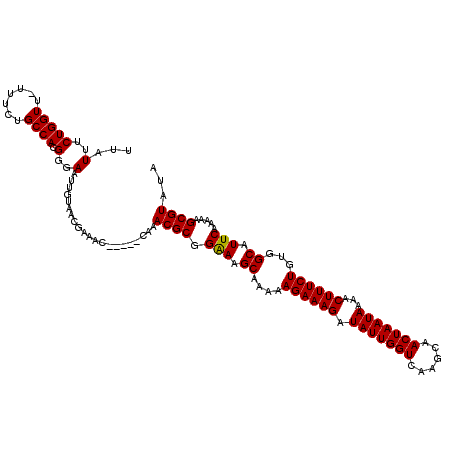
| Location | 3,393,736 – 3,393,847 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -22.39 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3393736 111 + 27905053 CAUACCAAACGCGGAAAGCAAAAAGAAAGAUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUAGCACUGAAUUCUGUUUUCACCUGA-----AAAAA-AA--- ........((((.(((.((....((((((.(((((((......)))))))...))))))...)).))).....))))................(((((....))-----)))..-..--- ( -21.30) >DroSim_CAF1 2652 116 + 1 CAAACCAAACGCGGAAAGCAAAAAGAAAGAUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUAGCACUGAAUCCUGUUUUCACCUGGAUGCCAAAAA-AA--- ..........((.....))....((((((.(((((((......)))))))...)))))).(((((((((....((.....))..((((.......))))..)))))))))....-..--- ( -26.70) >DroEre_CAF1 812 115 + 1 -----CAAACGCGGGAAGCAAAAAGAAAGGUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUAGUAGUGAAUCCUGUUUUCACCUGGUUACCAAAAAAAAAAG -----...((((..(((((....((((((.(((((((......)))))))...))))))...)).))).....))))......(((((.......))))).(((...))).......... ( -25.30) >DroYak_CAF1 6321 112 + 1 -----CAAACGCGGGAAGCAAAAAGAAAGGUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUAGUACUGAAUCCUGUUUUCACCUGGUUCCCAAAAAAAA--- -----.......(((((.((...((((((.(((((((......)))))))...))))))..((.(((((....((.....))..)))))))..........)).)))))........--- ( -26.20) >consensus _____CAAACGCGGAAAGCAAAAAGAAAGAUAUUGGUCAAGCAACUAAUAAAACUUUCUGUGGCAUUCAAAAAGCGUAUAGCACUGAAUCCUGUUUUCACCUGGUU_CCAAAAA_AA___ ..........(..(((((((...((((((.(((((((......)))))))...))))))..((.(((((....((.....))..))))))))))))))..)................... (-22.39 = -21.70 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:46 2006