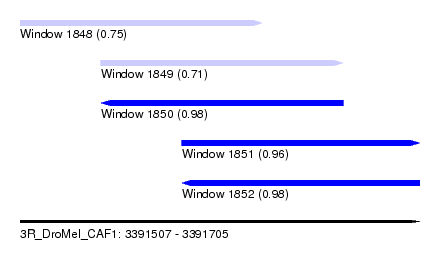
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,391,507 – 3,391,705 |
| Length | 198 |
| Max. P | 0.984603 |

| Location | 3,391,507 – 3,391,627 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -25.63 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3391507 120 + 27905053 UACUUACAUAUUUAGAUUUCUGGAUAUUUUAAGUGUACGCUUUAUGGUCAUGCAGAAGGUUGGAUGAAUUCGUUUCUCGCCUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCU ((((((.((((((((....))))))))..))))))..((((...((((...((.((((((.(((..........))).)))))))).(((....))).))))....)))).......... ( -28.90) >DroSim_CAF1 368 120 + 1 UACUUAUAUAUUAAGAUUUCUGCAUAUUUUAAGUGUACGCAUUAUGGUCAUGCAGAAGGUUGGAUGAAUUUGUUUCUCGCCUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCU ..((((.....))))..(((((((((((...((((....))))..))).))))))))((.(((...((((((......((....)).(((....)))...))))))....)))....)). ( -24.20) >DroYak_CAF1 4081 120 + 1 UACUUUUAUAUUUAGAGUUCUAAAUAUUUUAAGUGUACGCAUUAUGUUCAUGCAGAAGGUUGGAUGAAUUUGUUUCUCGCCUUCGCUGUCUAAUGACAAUCAAAUUAGCGGCAUUUUCCU (((((..((((((((....))))))))...)))))...((((.......)))).((((((.(((..........))).))))))((((.(((((.........)))))))))........ ( -31.40) >consensus UACUUAUAUAUUUAGAUUUCUGAAUAUUUUAAGUGUACGCAUUAUGGUCAUGCAGAAGGUUGGAUGAAUUUGUUUCUCGCCUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCU (((((..((((((((....))))))))...)))))........(((((...((.((((((.(((..........))).))))))...(((....)))..........)))))))...... (-25.63 = -25.63 + 0.00)

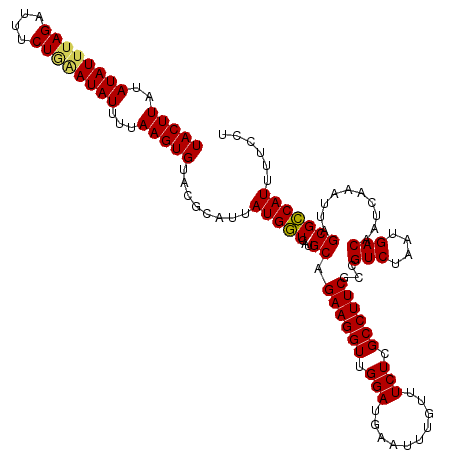
| Location | 3,391,547 – 3,391,667 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.81 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3391547 120 + 27905053 UUUAUGGUCAUGCAGAAGGUUGGAUGAAUUCGUUUCUCGCCUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUC ..........((((((((((.(((..........))).))))))..(((.(((((............((((((.......)))))).............))))).)))))))........ ( -29.61) >DroSim_CAF1 408 120 + 1 AUUAUGGUCAUGCAGAAGGUUGGAUGAAUUUGUUUCUCGCCUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUC ..........((((((((((.(((..........))).))))))..(((.(((((............((((((.......)))))).............))))).)))))))........ ( -29.61) >DroYak_CAF1 4121 120 + 1 AUUAUGUUCAUGCAGAAGGUUGGAUGAAUUUGUUUCUCGCCUUCGCUGUCUAAUGACAAUCAAAUUAGCGGCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUC ..........((((((((((.(((..........))).))))))..((((....)))).........(((.((.......)).)))......................))))........ ( -23.90) >consensus AUUAUGGUCAUGCAGAAGGUUGGAUGAAUUUGUUUCUCGCCUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUC ..........((((((((((.(((..........))).))))))..(((.(((((............((((((.......)))))).............))))).)))))))........ (-26.70 = -26.81 + 0.11)

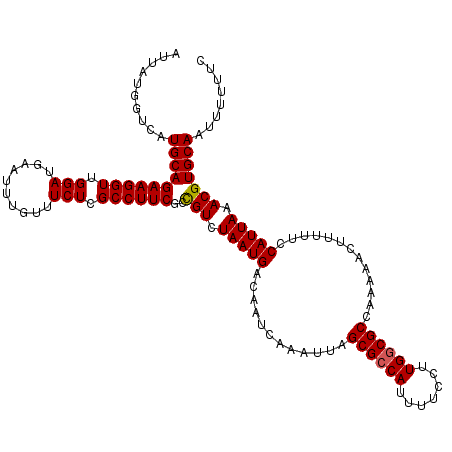
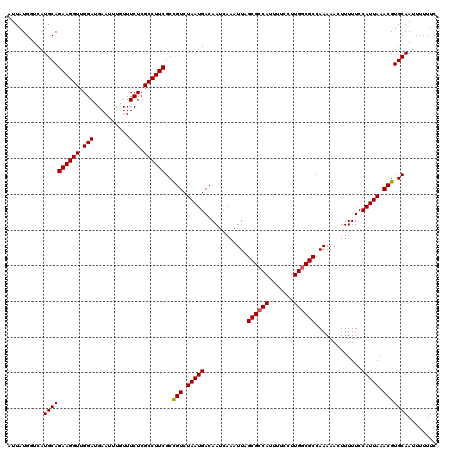
| Location | 3,391,547 – 3,391,667 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -31.43 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3391547 120 - 27905053 GAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGGCGCUAAUUUGAUUGUCAUUAGACGGCGAAGGCGAGAAACGAAUUCAUCCAACCUUCUGCAUGACCAUAAA ........((((((((((((((....((((.(((((((((.......)))))))))...)))).))))))))))..(((((.(((((.....))).))...))))))))).......... ( -35.90) >DroSim_CAF1 408 120 - 1 GAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGGCGCUAAUUUGAUUGUCAUUAGACGGCGAAGGCGAGAAACAAAUUCAUCCAACCUUCUGCAUGACCAUAAU ........((((((((((((((....((((.(((((((((.......)))))))))...)))).))))))))))..(((((.(((((.....))).))...))))))))).......... ( -35.90) >DroYak_CAF1 4121 120 - 1 GAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGCCGCUAAUUUGAUUGUCAUUAGACAGCGAAGGCGAGAAACAAAUUCAUCCAACCUUCUGCAUGAACAUAAU ........((((.(((((((((....((((.((((((.((.......)).))))))...)))).)))))))))...(((((.(((((.....))).))...))))))))).......... ( -26.60) >consensus GAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGGCGCUAAUUUGAUUGUCAUUAGACGGCGAAGGCGAGAAACAAAUUCAUCCAACCUUCUGCAUGACCAUAAU ........((((((((((((((....((((.(((((((((.......)))))))))...)))).))))))))))..(((((.(((((.....))).))...))))))))).......... (-31.43 = -32.10 + 0.67)

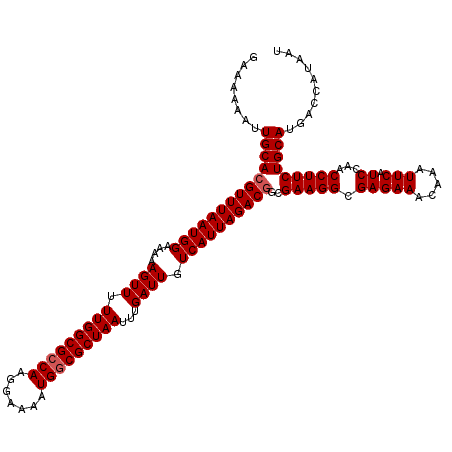
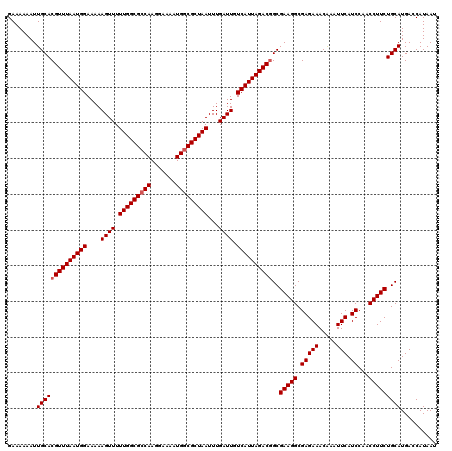
| Location | 3,391,587 – 3,391,705 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -16.74 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3391587 118 + 27905053 CUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUCUUUGUAUACAUUUUU--UUUUUAACUUGCCUCAAUAAAUU ....(((((.(((((............((((((.......)))))).............))))).))).))........................--....................... ( -19.01) >DroSim_CAF1 448 120 + 1 CUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUCUUUGUAUACAUUUUUAUUUUUUAACUUGCCUCAAUAAAUU ....(((((.(((((............((((((.......)))))).............))))).))).))................................................. ( -19.01) >DroYak_CAF1 4161 111 + 1 CUUCGCUGUCUAAUGACAAUCAAAUUAGCGGCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUCUUCAUUUA--UUUUU--UUU-----UUCCCUCAAUAAAUU ....(((((.(((((............(((.((.......)).))).............))))).))).)).................--.....--...-----............... ( -12.21) >consensus CUUCGCCGUCUAAUGACAAUCAAAUUAGCGCCAUUUUCCUUGGCGCCAAAAACUUUUUCCAUUAAACGUGCAAUUUUUUCUUUGUAUACAUUUUU__UUUUUAACUUGCCUCAAUAAAUU ....(((((.(((((............((((((.......)))))).............))))).))).))................................................. (-15.93 = -16.04 + 0.11)

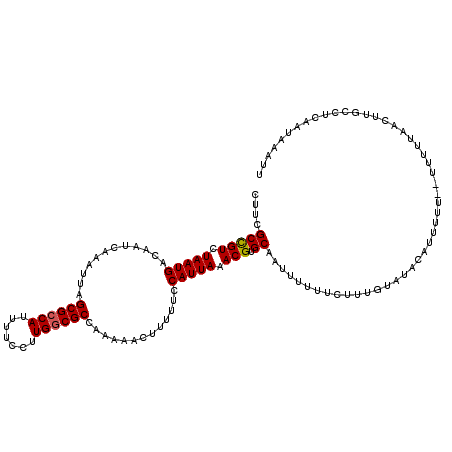
| Location | 3,391,587 – 3,391,705 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
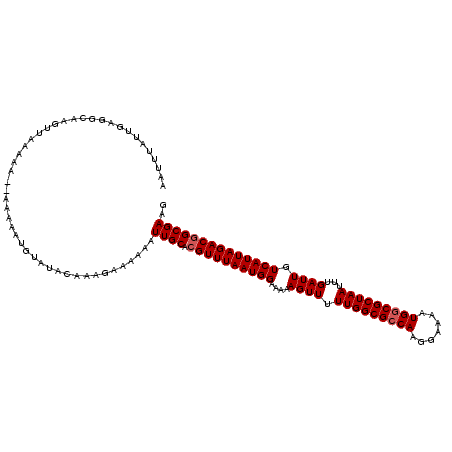
>3R_DroMel_CAF1 3391587 118 - 27905053 AAUUUAUUGAGGCAAGUUAAAAA--AAAAAUGUAUACAAAGAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGGCGCUAAUUUGAUUGUCAUUAGACGGCGAAG ..(((.(((..(((..((.....--.))..)))...))).)))....((((.((((((((((....((((.(((((((((.......)))))))))...)))).)))))))))))))).. ( -32.00) >DroSim_CAF1 448 120 - 1 AAUUUAUUGAGGCAAGUUAAAAAAUAAAAAUGUAUACAAAGAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGGCGCUAAUUUGAUUGUCAUUAGACGGCGAAG ..(((.(((..(((..((........))..)))...))).)))....((((.((((((((((....((((.(((((((((.......)))))))))...)))).)))))))))))))).. ( -31.80) >DroYak_CAF1 4161 111 - 1 AAUUUAUUGAGGGAA-----AAA--AAAAA--UAAAUGAAGAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGCCGCUAAUUUGAUUGUCAUUAGACAGCGAAG .(((((((.......-----...--...))--)))))..........((((..(((((((((....((((.((((((.((.......)).))))))...)))).))))))))).)))).. ( -20.82) >consensus AAUUUAUUGAGGCAAGUUAAAAA__AAAAAUGUAUACAAAGAAAAAAUUGCACGUUUAAUGGAAAAAGUUUUUGGCGCCAAGGAAAAUGGCGCUAAUUUGAUUGUCAUUAGACGGCGAAG ...............................................((((.((((((((((....((((.(((((((((.......)))))))))...)))).)))))))))))))).. (-25.90 = -26.57 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:42 2006