
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,383,210 – 3,383,308 |
| Length | 98 |
| Max. P | 0.945930 |

| Location | 3,383,210 – 3,383,308 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.06 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
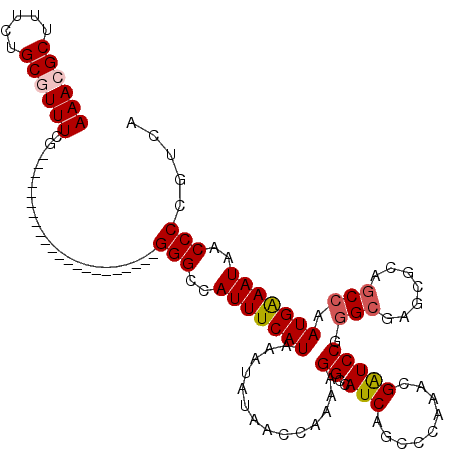
>3R_DroMel_CAF1 3383210 98 + 27905053 AAACGCUUUCUGCGUUUCG----------------------GGGCCAUUUCAUAAAUAUAACCAAAAGGCCAUCAGUACAAACGAUCCGGGCGAGCGCAGCCAAUGAAAUAACCCCGUCA ((((((.....))))))((----------------------(((..(((((((..............((..(((.((....))))))).(((.......))).)))))))..)))))... ( -27.10) >DroSec_CAF1 11583 98 + 1 AAACGCUUUCUGCGUUUCG----------------------GGGCCAUUUCAUAAAUAUAACCAAAAGGCCAUCAGCCCAAACGAUCCGGGCGAGCGCAGCCAAUGAAAUAACCCCGUCA ((((((.....))))))((----------------------(((..(((((((..............(((.....)))...........(((.......))).)))))))..)))))... ( -29.10) >DroAna_CAF1 19959 120 + 1 AAACGCUUUUUGCCUUUCGUGGACAUUUGAUAGUCCGAGUUGGGCAAUUUCAUAAAUAUAACGAGAAGGCCAUCAGAGGGAACGGUCCAGGCAAGCGAAACCAAUGGAAUGACCCCAUCG ...((((...(((((.((((((((........))))((....(((..((((...........))))..))).)).......))))...)))))))))......((((.......)))).. ( -29.80) >DroSim_CAF1 12418 98 + 1 AAACGCUUUCUGCGUUUCG----------------------GGGCCAUUUCAUAAAUAUAACCAAAAGGCCAUCAGCCCAAACGAUCCGGGCGAGCGCAGCCAAUGAAAUAACCCCGUCA ((((((.....))))))((----------------------(((..(((((((..............(((.....)))...........(((.......))).)))))))..)))))... ( -29.10) >DroEre_CAF1 11812 97 + 1 AAACGCUUUCUGCCUUUCG----------------------GGGCCAUUUCAUAAAUAUAACCAAAAGGCCAUCAGCCCAAACGAUCCG-GCGAGCGCAGCCAAUGAAAUAACCCGGACA ....((.....))..((((----------------------((...(((((((..............(((.....)))..........(-((.......))).)))))))..)))))).. ( -23.50) >consensus AAACGCUUUCUGCGUUUCG______________________GGGCCAUUUCAUAAAUAUAACCAAAAGGCCAUCAGCCCAAACGAUCCGGGCGAGCGCAGCCAAUGAAAUAACCCCGUCA ((((((.....))))))........................(((..(((((((..............((..(((.........))))).(((.......))).)))))))..)))..... (-16.58 = -17.06 + 0.48)

| Location | 3,383,210 – 3,383,308 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -23.50 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3383210 98 - 27905053 UGACGGGGUUAUUUCAUUGGCUGCGCUCGCCCGGAUCGUUUGUACUGAUGGCCUUUUGGUUAUAUUUAUGAAAUGGCCC----------------------CGAAACGCAGAAAGCGUUU ...((((((((((((((..((.(((.((.....)).)))..))....((((((....))))))....))))))))))))----------------------))((((((.....)))))) ( -35.90) >DroSec_CAF1 11583 98 - 1 UGACGGGGUUAUUUCAUUGGCUGCGCUCGCCCGGAUCGUUUGGGCUGAUGGCCUUUUGGUUAUAUUUAUGAAAUGGCCC----------------------CGAAACGCAGAAAGCGUUU ...((((((((((((((...........(((((((...)))))))..((((((....))))))....))))))))))))----------------------))((((((.....)))))) ( -41.10) >DroAna_CAF1 19959 120 - 1 CGAUGGGGUCAUUCCAUUGGUUUCGCUUGCCUGGACCGUUCCCUCUGAUGGCCUUCUCGUUAUAUUUAUGAAAUUGCCCAACUCGGACUAUCAAAUGUCCACGAAAGGCAAAAAGCGUUU (((((((.....)))))))....(((((((((((.(((((......)))))))...((((........((........))....((((........)))))))).)))))...))))... ( -31.30) >DroSim_CAF1 12418 98 - 1 UGACGGGGUUAUUUCAUUGGCUGCGCUCGCCCGGAUCGUUUGGGCUGAUGGCCUUUUGGUUAUAUUUAUGAAAUGGCCC----------------------CGAAACGCAGAAAGCGUUU ...((((((((((((((...........(((((((...)))))))..((((((....))))))....))))))))))))----------------------))((((((.....)))))) ( -41.10) >DroEre_CAF1 11812 97 - 1 UGUCCGGGUUAUUUCAUUGGCUGCGCUCGC-CGGAUCGUUUGGGCUGAUGGCCUUUUGGUUAUAUUUAUGAAAUGGCCC----------------------CGAAAGGCAGAAAGCGUUU ..((.((((((((((((((((.......))-))(((((...((((.....))))..)))))......))))))))))))----------------------.))((.((.....)).)). ( -30.90) >consensus UGACGGGGUUAUUUCAUUGGCUGCGCUCGCCCGGAUCGUUUGGGCUGAUGGCCUUUUGGUUAUAUUUAUGAAAUGGCCC______________________CGAAACGCAGAAAGCGUUU .....((((((((((((.(((.......))).((.(((((......)))))))..............))))))))))))........................((((((.....)))))) (-23.50 = -24.18 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:30 2006