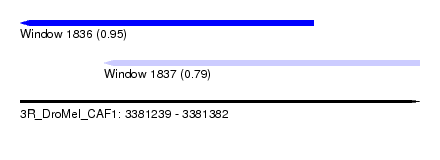
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,381,239 – 3,381,382 |
| Length | 143 |
| Max. P | 0.953824 |

| Location | 3,381,239 – 3,381,344 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -21.62 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
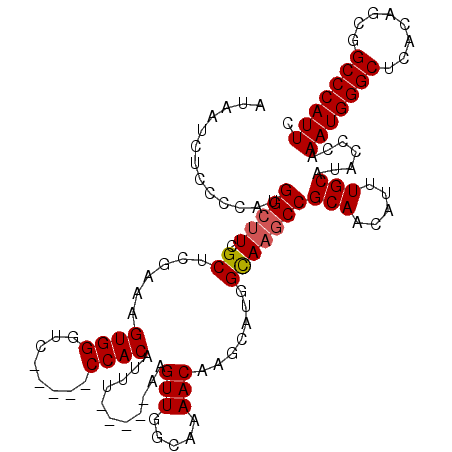
>3R_DroMel_CAF1 3381239 105 - 27905053 CACAUUU-----AAGUUGGCAAAACAAGCAUGGCAUGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCCGCCCAUUCUUUCAUGCCAAAAA-U-AAAACGGCCCUUGUA ......(-----(((..(((..........(((((((..(((.....))).......(((((((........)))))))....)))))))....-.-......))))))).. ( -26.80) >DroSec_CAF1 9573 107 - 1 CACAUUU-----AAGUUGGCAAAACAUGCAUGGCAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUCUUCCAUGCCAAAAAAUAAAAACGGCCCUUGUA .(((...-----..(((((((.....))).(((((....(((.....))).......((((((((......))))))))......)))))...........))))...))). ( -25.50) >DroSim_CAF1 10410 107 - 1 CACAUUU-----AAGUUGGCAAAACAAGCAUCGUAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUCUUCCAUGCCAAAAAAUAAAAACGGCCCUUGUA ......(-----(((..(((.......((..(....)..)).......((((.....((((((((......)))))))).....))))...............))))))).. ( -22.10) >DroEre_CAF1 9925 110 - 1 CACAUUUAACAUAAGUUGGCAAAACAAGCAUGGCAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUCUUCCAUGCCAAAAA-U-AAAAAGCCCGUUGUA ......((((....(((.....)))..((((((.(((..(((.....))).......((((((((......)))))))))))))))))......-.-.........)))).. ( -26.10) >consensus CACAUUU_____AAGUUGGCAAAACAAGCAUGGCAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUCUUCCAUGCCAAAAA_U_AAAACGGCCCUUGUA ...............((((((....(((.((((...(((((..((((((......))))))(.....).))))))))).)))...))))))..................... (-21.62 = -22.38 + 0.75)


| Location | 3,381,269 – 3,381,382 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -24.71 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3381269 113 - 27905053 AUAACCUCCCCAUGGCUUCGCUCGAAAGUGGGCUUC--GCCACAUUU-----AAGUUGGCAAAACAAGCAUGGCAUGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCCGCCCAUUC .........(((((.(((.(((((....)))))...--(((((....-----..).)))).....))))))))......(((.....))).......(((((((........))))))). ( -29.10) >DroSec_CAF1 9605 110 - 1 AUAAUCUUCCCAUGGCUUCGCUCGAAAGUGGGUC-----CCACAUUU-----AAGUUGGCAAAACAUGCAUGGCAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUC .............(((((.(((.....((((...-----))))....-----......(((.....)))..))))))))(((.....))).......((((((((......)))))))). ( -29.10) >DroSim_CAF1 10442 110 - 1 AUAAUCUCCCCAUGGCUUCGCUCGAAAGUGGGUC-----CCACAUUU-----AAGUUGGCAAAACAAGCAUCGUAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUC .............(((((((((..((.((((...-----)))).)).-----.)))..((.......))...).)))))(((.....))).......((((((((......)))))))). ( -27.80) >DroEre_CAF1 9955 117 - 1 AUAAG---CCCUUGGGUUUGCUCAAAAGUGGCUCCCAUCCCACAUUUAACAUAAGUUGGCAAAACAAGCAUGGCAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUC .....---....(((((.(((......((((((.((((........((((....))))((.......))))))..)))))).......))).)))))((((((((......)))))))). ( -36.32) >consensus AUAAUCUCCCCAUGGCUUCGCUCGAAAGUGGGUC_____CCACAUUU_____AAGUUGGCAAAACAAGCAUGGCAAGCCGCAACAUUUGCAUACCCAAAUGGGCUCACAGCGGCCCAUUC .............(((((.((......((((........))))...........(((.....))).......)))))))(((.....))).......(((((((........))))))). (-24.71 = -25.02 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:28 2006