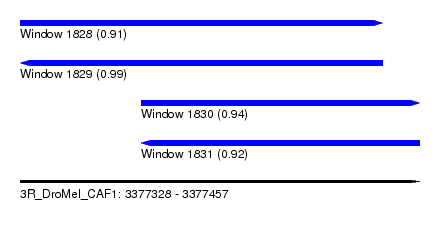
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,377,328 – 3,377,457 |
| Length | 129 |
| Max. P | 0.988246 |

| Location | 3,377,328 – 3,377,445 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -20.55 |
| Energy contribution | -23.15 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3377328 117 + 27905053 CC-CGGGGCUGAGAUUGGGUUAAGCAAGGGGAAGCCCCUAAAGAGCGAAGGUAAGGUAAGUUUAAGGGCACUUUUCCCGUUCAAGGGGA--AAAGGUGCAAACCCAAAACUUAACUUUCC ..-.((((((....(((.......))).....))))))........((((((....(((((((..(((((((((((((.......))))--))).))))...))..))))))))))))). ( -37.90) >DroSec_CAF1 5704 117 + 1 CC-AGGGGCUGAGAUUGGGUUAAGCAAGGGGAAGACCCCAAAGAGCGAAGGUAAGGGAAGUUUAAGGGCACUUUUCCCGUUCCAGGGGA--AAAGGUGCAAACCCUAAACUUAACUUUCC ..-.((((((....(((.......))).....)).))))..........((.(((..(((((((.(((((((((((((.......))))--))).))))...)).)))))))..))).)) ( -36.90) >DroAna_CAF1 14865 96 + 1 CCUAGGGCCUCAGAUUGGGUUAA------------------------AAGGUAAAAAGAGUUAAGAGUCACUUCUCCUGUUCAGGAGGAAAAAAGGUUCAAGCCAUAAAGUCAGCUUUUA (((..(((((......)))))..------------------------.)))....(((((((.........(((((((....))))))).....(((....)))........))))))). ( -22.20) >DroSim_CAF1 6542 116 + 1 CC-AGGGGCUGAGAUUGGGUUAAGCAAGG-GAAGCCCCCAAAGAGCGAAGGUAAGGGAAGUUUAAGGGCACUUUUCCCGUUCAAGGGGA--AAAGGUGCAAACCCUAAACUUAACUUUCC ..-.((((((....(((.......)))..-..))))))...........((.(((..(((((((.(((((((((((((.......))))--))).))))...)).)))))))..))).)) ( -40.40) >DroEre_CAF1 6210 117 + 1 CC-AGGGGCUGAGAUUGGGUUAAGCUAGGGGAAGCCCCUGAAGAGCGAAGGUGAGGGAAGUUUAAGGGCCCUUUUGCCGUUCACGGGGA--AAAGGUGCAAGCCAUAAACUUAACUUUCU .(-(((((((....((((......))))....))))))))......((((((.....(((((((.((((((((((.((.......)).)--))))).))...)).))))))).)))))). ( -44.00) >consensus CC_AGGGGCUGAGAUUGGGUUAAGCAAGGGGAAGCCCCCAAAGAGCGAAGGUAAGGGAAGUUUAAGGGCACUUUUCCCGUUCAAGGGGA__AAAGGUGCAAACCCUAAACUUAACUUUCC ....((((((......................))))))........((((((.....(((((((.(((((((((((((.......))))...)))))))...)).))))))).)))))). (-20.55 = -23.15 + 2.60)

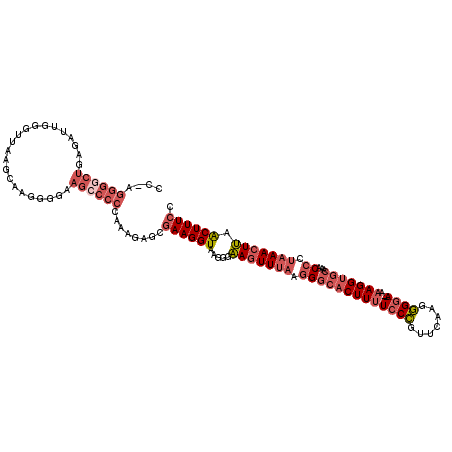
| Location | 3,377,328 – 3,377,445 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -33.21 |
| Consensus MFE | -17.61 |
| Energy contribution | -20.49 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3377328 117 - 27905053 GGAAAGUUAAGUUUUGGGUUUGCACCUUU--UCCCCUUGAACGGGAAAAGUGCCCUUAAACUUACCUUACCUUCGCUCUUUAGGGGCUUCCCCUUGCUUAACCCAAUCUCAGCCCCG-GG ((.(((.(((((((.(((...((((.(((--((((.......)))))))))))))).))))))).))).))...........((((((.....(((.......)))....)))))).-.. ( -37.60) >DroSec_CAF1 5704 117 - 1 GGAAAGUUAAGUUUAGGGUUUGCACCUUU--UCCCCUGGAACGGGAAAAGUGCCCUUAAACUUCCCUUACCUUCGCUCUUUGGGGUCUUCCCCUUGCUUAACCCAAUCUCAGCCCCU-GG ((.(((..((((((((((...((((.(((--((((.......)))))))))))))))))))))..))).))...((.....((((....))))..))....................-.. ( -38.00) >DroAna_CAF1 14865 96 - 1 UAAAAGCUGACUUUAUGGCUUGAACCUUUUUUCCUCCUGAACAGGAGAAGUGACUCUUAACUCUUUUUACCUU------------------------UUAACCCAAUCUGAGGCCCUAGG .....(((.......(((.(((((.........(((((....))))).(((........)))..........)------------------------)))).)))......)))...... ( -13.82) >DroSim_CAF1 6542 116 - 1 GGAAAGUUAAGUUUAGGGUUUGCACCUUU--UCCCCUUGAACGGGAAAAGUGCCCUUAAACUUCCCUUACCUUCGCUCUUUGGGGGCUUC-CCUUGCUUAACCCAAUCUCAGCCCCU-GG ((.(((..((((((((((...((((.(((--((((.......)))))))))))))))))))))..))).))........(..((((((..-..(((.......)))....)))))).-.) ( -42.30) >DroEre_CAF1 6210 117 - 1 AGAAAGUUAAGUUUAUGGCUUGCACCUUU--UCCCCGUGAACGGCAAAAGGGCCCUUAAACUUCCCUCACCUUCGCUCUUCAGGGGCUUCCCCUAGCUUAACCCAAUCUCAGCCCCU-GG .(((((..(((((((.((...((.(((((--(..(((....))).)))))))))).)))))))..))....)))......((((((((......................)))))))-). ( -34.35) >consensus GGAAAGUUAAGUUUAGGGUUUGCACCUUU__UCCCCUUGAACGGGAAAAGUGCCCUUAAACUUCCCUUACCUUCGCUCUUUAGGGGCUUCCCCUUGCUUAACCCAAUCUCAGCCCCU_GG ((.(((..((((((((((...((((.(((..((((.......)))))))))))))))))))))..))).))...........((((((......................)))))).... (-17.61 = -20.49 + 2.88)


| Location | 3,377,367 – 3,377,457 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3377367 90 + 27905053 AAGAGCGAAGGUAAGGUAAGUUUAAGGGCACUUUUCCCGUUCAAGGGGAAAAGGUGCAAACCCAAAACUUAACUUUCC---------UUCUUUU-GCGAG ....(((((((.((((.(((((.(((.(((((((((((.......))))))).))))..........)))))))).))---------)))))))-))... ( -32.00) >DroSec_CAF1 5743 90 + 1 AAGAGCGAAGGUAAGGGAAGUUUAAGGGCACUUUUCCCGUUCCAGGGGAAAAGGUGCAAACCCUAAACUUAACUUUCC---------UUCUUUU-GGGAG ......(((((.(((..(((((((.(((((((((((((.......))))))).))))...)).)))))))..))).))---------)))....-..... ( -33.70) >DroSim_CAF1 6580 90 + 1 AAGAGCGAAGGUAAGGGAAGUUUAAGGGCACUUUUCCCGUUCAAGGGGAAAAGGUGCAAACCCUAAACUUAACUUUCC---------UUCUUUU-GGGAG ......(((((.(((..(((((((.(((((((((((((.......))))))).))))...)).)))))))..))).))---------)))....-..... ( -33.70) >DroEre_CAF1 6249 100 + 1 AAGAGCGAAGGUGAGGGAAGUUUAAGGGCCCUUUUGCCGUUCACGGGGAAAAGGUGCAAGCCAUAAACUUAACUUUCUCUGGAUACAUUCUUUUCGGCAG ....((((((((.....(((((((.((((((((((.((.......)).)))))).))...)).))))))).))))))...(((.....))).....)).. ( -27.00) >consensus AAGAGCGAAGGUAAGGGAAGUUUAAGGGCACUUUUCCCGUUCAAGGGGAAAAGGUGCAAACCCUAAACUUAACUUUCC_________UUCUUUU_GGGAG .........((.(((..(((((((.(((((((((((((.......))))))).))))...)).)))))))..))).))...................... (-22.80 = -23.18 + 0.38)


| Location | 3,377,367 – 3,377,457 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -22.62 |
| Energy contribution | -24.12 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.73 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3377367 90 - 27905053 CUCGC-AAAAGAA---------GGAAAGUUAAGUUUUGGGUUUGCACCUUUUCCCCUUGAACGGGAAAAGUGCCCUUAAACUUACCUUACCUUCGCUCUU .....-....(((---------((.(((.(((((((.(((...((((.(((((((.......)))))))))))))).))))))).))).)))))...... ( -32.90) >DroSec_CAF1 5743 90 - 1 CUCCC-AAAAGAA---------GGAAAGUUAAGUUUAGGGUUUGCACCUUUUCCCCUGGAACGGGAAAAGUGCCCUUAAACUUCCCUUACCUUCGCUCUU .....-....(((---------((.(((..((((((((((...((((.(((((((.......)))))))))))))))))))))..))).)))))...... ( -35.70) >DroSim_CAF1 6580 90 - 1 CUCCC-AAAAGAA---------GGAAAGUUAAGUUUAGGGUUUGCACCUUUUCCCCUUGAACGGGAAAAGUGCCCUUAAACUUCCCUUACCUUCGCUCUU .....-....(((---------((.(((..((((((((((...((((.(((((((.......)))))))))))))))))))))..))).)))))...... ( -35.70) >DroEre_CAF1 6249 100 - 1 CUGCCGAAAAGAAUGUAUCCAGAGAAAGUUAAGUUUAUGGCUUGCACCUUUUCCCCGUGAACGGCAAAAGGGCCCUUAAACUUCCCUCACCUUCGCUCUU ....................((((..((..(((((((.((...((.((((((..(((....))).)))))))))).)))))))..))........)))). ( -21.40) >consensus CUCCC_AAAAGAA_________GGAAAGUUAAGUUUAGGGUUUGCACCUUUUCCCCUUGAACGGGAAAAGUGCCCUUAAACUUCCCUUACCUUCGCUCUU ......................((.(((..((((((((((...((((.(((((((.......)))))))))))))))))))))..))).))......... (-22.62 = -24.12 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:22 2006