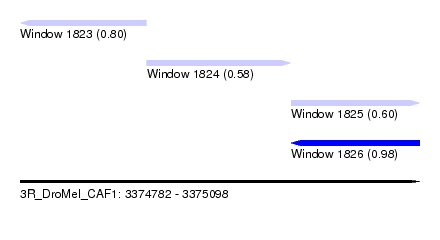
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,374,782 – 3,375,098 |
| Length | 316 |
| Max. P | 0.982950 |

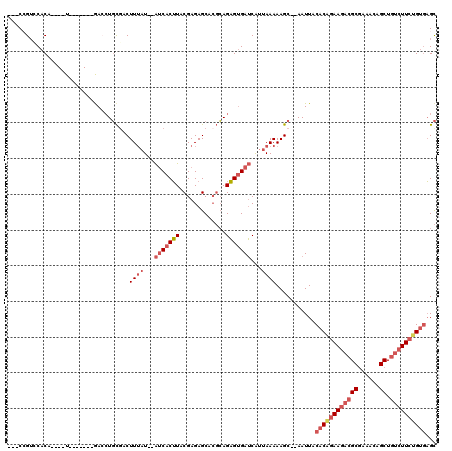
| Location | 3,374,782 – 3,374,882 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -26.04 |
| Energy contribution | -27.36 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3374782 100 - 27905053 AGCCGCUACUGAAUCGAGGCUGCUGCGGG-CUU----A--C-----UGGCAGUCGAACUGAUGGAUCCGAUUGUUCGGUUCU--------CAUUGCGCGAUUCAGUUGUUCCUCGUCGAU ....((.(((((((((..((.(((((.((-...----.--)-----).)))))......((((((.((((....)))).)).--------)))))).))))))))).))........... ( -34.00) >DroSec_CAF1 3173 102 - 1 AGCGGCAACUGAAUCGAGGCUGCUGCGGG-CUU----ACAC-----UGGCAGUCGAACUGAUGGAUCCGAUUGUUCGGUUCU--------CAUUGCGCGAUUCAGUUGUUCCUCGUCGAU ((..((((((((((((..((.(((((.((-...----...)-----).)))))......((((((.((((....)))).)).--------)))))).))))))))))))..))....... ( -40.00) >DroAna_CAF1 7931 118 - 1 AACCGCAACUGAAUUGGGGCUGCUGUGGGGCUUGCUUGCACCGCUCCGCCACUCGAACUGAUAGAUCCGAUUCUACG-UUCUCGGUGUUCCAGUGCC-CAGUCAGUUGUUCCUCGUCGAU ....((((((((.(((((((((..(((((((.((....))..)))))))((((.((((.(.((((......))))))-)))..))))...)))).))-)))))))))))........... ( -41.10) >DroSim_CAF1 4000 102 - 1 AGCGGCAACUGAAUCGAGGCUGCUGCGGG-CUU----ACAC-----UGGCAGUCGAACUGAUGGAUCCGAUUGUUCGGUUCU--------CAUUGCGCGAUUCAGUUGUUCCUCGUCGAU ((..((((((((((((..((.(((((.((-...----...)-----).)))))......((((((.((((....)))).)).--------)))))).))))))))))))..))....... ( -40.00) >DroEre_CAF1 3448 99 - 1 AGCCACAACUGAAUUGAGGCUGCUGUGGG-CUC------AC-----UGGCAGUCGAACUGAUGGAUCGGAUUGUUCGGUUC---------CAUUGCACGAUUCAGUUGUUCCUCGUCAAU ....((((((((((((.((((((((((..-..)------))-----.))))))).....((((((.(.((....)).).))---------))))...))))))))))))........... ( -37.60) >consensus AGCCGCAACUGAAUCGAGGCUGCUGCGGG_CUU____ACAC_____UGGCAGUCGAACUGAUGGAUCCGAUUGUUCGGUUCU________CAUUGCGCGAUUCAGUUGUUCCUCGUCGAU ....((((((((((((.(((((((.......................)))))))(((((((.............)))))))................))))))))))))........... (-26.04 = -27.36 + 1.32)

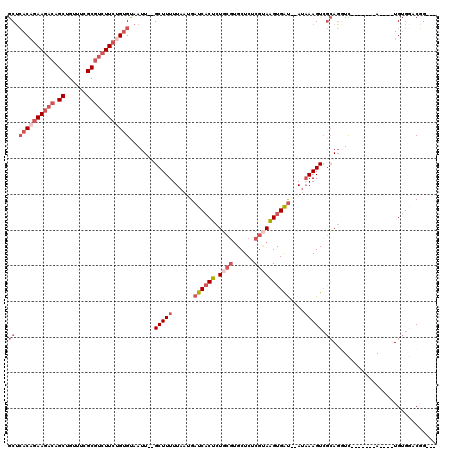
| Location | 3,374,882 – 3,374,996 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -38.84 |
| Consensus MFE | -23.28 |
| Energy contribution | -25.12 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3374882 114 + 27905053 UUAAUCACGACCGCCGCUUAUCGCAAUGUGAGUUGCUGCUAUCGCU---GCAGCGAAACAUAUGGCGCAGAAUAUUGGAGUAGAAUAUCGCGACUGAUAAGCGGAGCGGACCGAAC---U .......((.((((((((((((((.(((((..(((((((.......---)))))))..))))).))((.((.((((.......))))))))....))))))))..))))..))...---. ( -39.40) >DroSec_CAF1 3275 114 + 1 UUAAUCACGACCGCCGCUUAUCGCAAUGUGAGUUGCUGC---CGCU---GCAGCGAAACAUAUGGCGCAGAAUAUUGGAGUAGAAUAUCGCGACUGAUAAGCGGAACGGGCCGAACCGCU .......((.((((((((((((((.(((((..(((((((---....---)))))))..))))).))((.((.((((.......))))))))....)))))))))..)))..))....... ( -40.00) >DroAna_CAF1 8049 109 + 1 UUAAUCACGCUCGCCGCUUAUCG-ACGCUGAGUUGAUGUUGUUGUUGGCGGUGCGAAACAUAUGGCCGGG------GAAGUAGAACAUCGGGACUGAUAAGCGGAGAGCUCUGAAC---- ....(((.((((.((((((((((-((.....)))((((((.((.(((((.(((.......))).))))).------)).....))))))......))))))))).))))..)))..---- ( -39.00) >DroSim_CAF1 4102 114 + 1 UUAAUCACGACCGCCGCUUAUCGCAAUGUGAGUUGCUGC---CGCU---GCAGCGAAACAUAUGGCGCAGAAUAUUGGAGUAGAAUAUCGCGACUGAUAAGCGGAGCGGGCCGAACUGCU .............(((((((((((.(((((..(((((((---....---)))))))..))))).))((.((.((((.......))))))))....)))))))))(((((......))))) ( -40.90) >DroEre_CAF1 3547 110 + 1 UUAAUCACGAUUGCCCCUUAUCGCAAUAUGGGUUGC-GC---CGCU---GCACCGAAACAUAUGGCGCAGAAUCUUGGAGCUGAAUAUCGCGACUGAUAAGCGGCGAGGGCAGAGC---U ..........((((((.((..(((..(((.((((((-(.---..((---((.(((.......))).))))..((........))....))))))).))).)))..))))))))...---. ( -34.90) >consensus UUAAUCACGACCGCCGCUUAUCGCAAUGUGAGUUGCUGC___CGCU___GCAGCGAAACAUAUGGCGCAGAAUAUUGGAGUAGAAUAUCGCGACUGAUAAGCGGAGCGGGCCGAAC___U ..........((.(((((((((((.(((((..(((((((..........)))))))..))))).))............(((.(.....)...))))))))))))...))........... (-23.28 = -25.12 + 1.84)

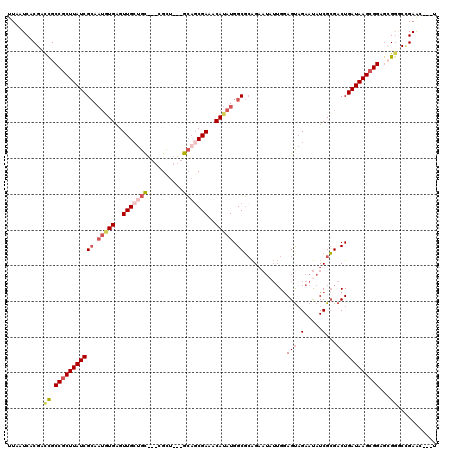
| Location | 3,374,996 – 3,375,098 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -17.18 |
| Energy contribution | -20.10 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3374996 102 + 27905053 GCUCACAGAAGACAGCUGUUUCGCGUCUUCUGGGUAAUU--GCUUUUUAAUGAUCACUCUGCGUGCUCUCGUAAGUGAU--AUAAAGUCGCAGGUC-------A----UGUGGACGG--- .....((((((((.((......)))))))))).....((--((.(((((...((((((.((((......))))))))))--.)))))..))))(((-------.----....)))..--- ( -30.00) >DroSec_CAF1 3389 102 + 1 GCUCACAGAAGACAGCUGUUUGGCGUCUUCUGUGUAAUU--GCUUUUUAAUGAUCACUCUGCGUGCUCUCGUAAGUGAU--AUAAAGUCGCAGGUU-------A----UGUGGACGG--- ((.((((((((((.(((....))))))))))))).....--)).........((((((.((((......))))))))))--.....((((((....-------.----))).)))..--- ( -33.50) >DroAna_CAF1 8158 119 + 1 ACAGACCGAAGACAGCUGUUUCGCCGGUUGUGUGUAAUUACGCUUUUUAACGCUC-CACUCACUGCUCUCGCAUGUGGCUCAAAAAGUCAGAUUUUCAGACUUAUCUUUGAGAUCUGUCA (((((.((...(((((((......)))))))((((....)))).......))..(-(((....(((....))).))))(((((((((((.(.....).)))))...)))))).))))).. ( -27.70) >DroSim_CAF1 4216 102 + 1 GCUCACAGAAGACAGCUGUUUCGCGUCUUCUGUGUAAUU--GCUUUUUAAUGAUCACUCUGCGUGCUCUCGUAAGUGAU--AUUAAGUAGCAGGUC-------A----UGUGGACGG--- ...((((((((((.((......))))))))))))...((--(((.(((((((.(((((.((((......))))))))))--)))))).)))))(((-------.----....)))..--- ( -37.70) >DroEre_CAF1 3657 102 + 1 GCUCACAGAAGACAGCUGUUUCGCGUCUUCCGUGUAAUU--GCUUUUUAAUGAUCACUCUGCGAGCUCUCGAAAGUGAU--AUAAAGUCGCAGUUC-------A----UGUGCACUG--- ((.....((((((.((......))))))))((((.((((--((.(((((...((((((...(((....)))..))))))--.)))))..)))))))-------)----)).))....--- ( -30.10) >consensus GCUCACAGAAGACAGCUGUUUCGCGUCUUCUGUGUAAUU__GCUUUUUAAUGAUCACUCUGCGUGCUCUCGUAAGUGAU__AUAAAGUCGCAGGUC_______A____UGUGGACGG___ ((.((((((((((.((......)))))))))))).......(((((......((((((.((((......))))))))))....))))).))............................. (-17.18 = -20.10 + 2.92)

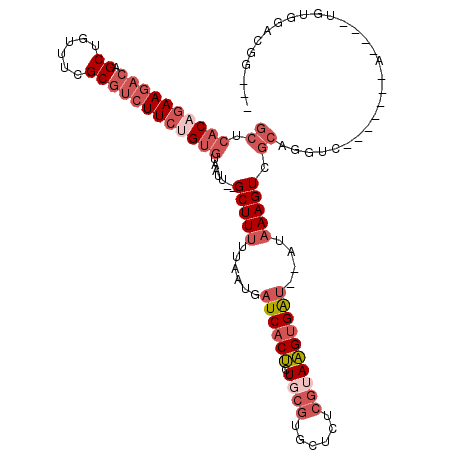
| Location | 3,374,996 – 3,375,098 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -15.76 |
| Energy contribution | -17.72 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3374996 102 - 27905053 ---CCGUCCACA----U-------GACCUGCGACUUUAU--AUCACUUACGAGAGCACGCAGAGUGAUCAUUAAAAAGC--AAUUACCCAGAAGACGCGAAACAGCUGUCUUCUGUGAGC ---..(..(((.----.-------....(((...((((.--(((((((.((......))..)))))))...))))..))--).......(((((((((......)).))))))))))..) ( -25.20) >DroSec_CAF1 3389 102 - 1 ---CCGUCCACA----U-------AACCUGCGACUUUAU--AUCACUUACGAGAGCACGCAGAGUGAUCAUUAAAAAGC--AAUUACACAGAAGACGCCAAACAGCUGUCUUCUGUGAGC ---.........----.-------....(((...((((.--(((((((.((......))..)))))))...))))..))--)....((((((((((((......)).))))))))))... ( -27.80) >DroAna_CAF1 8158 119 - 1 UGACAGAUCUCAAAGAUAAGUCUGAAAAUCUGACUUUUUGAGCCACAUGCGAGAGCAGUGAGUG-GAGCGUUAAAAAGCGUAAUUACACACAACCGGCGAAACAGCUGUCUUCGGUCUGU ..((((((((((((...(((((.(.....).))))))))))(((...(((....)))(((.(((-(.(((((....)))))..)))).)))....)))(((((....)).)))))))))) ( -34.30) >DroSim_CAF1 4216 102 - 1 ---CCGUCCACA----U-------GACCUGCUACUUAAU--AUCACUUACGAGAGCACGCAGAGUGAUCAUUAAAAAGC--AAUUACACAGAAGACGCGAAACAGCUGUCUUCUGUGAGC ---..(((....----.-------))).((((..(((((--(((((((.((......))..))))))).)))))..)))--)....((((((((((((......)).))))))))))... ( -33.30) >DroEre_CAF1 3657 102 - 1 ---CAGUGCACA----U-------GAACUGCGACUUUAU--AUCACUUUCGAGAGCUCGCAGAGUGAUCAUUAAAAAGC--AAUUACACGGAAGACGCGAAACAGCUGUCUUCUGUGAGC ---....((...----.-------....(((...((((.--(((((((((((....))).))))))))...))))..))--)....((((((((((((......)).)))))))))).)) ( -33.40) >consensus ___CCGUCCACA____U_______GACCUGCGACUUUAU__AUCACUUACGAGAGCACGCAGAGUGAUCAUUAAAAAGC__AAUUACACAGAAGACGCGAAACAGCUGUCUUCUGUGAGC ..................................((((...(((((((.............)))))))...))))...........((((((((((((......)).))))))))))... (-15.76 = -17.72 + 1.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:18 2006