| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,341,327 – 3,341,423 |
| Length | 96 |
| Max. P | 0.921731 |

| Location | 3,341,327 – 3,341,423 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -16.62 |
| Energy contribution | -15.57 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
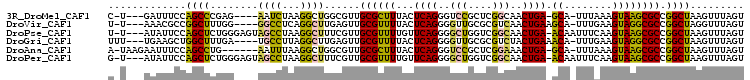
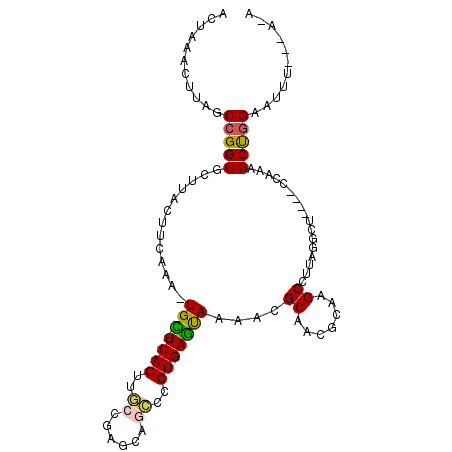
>3R_DroMel_CAF1 3341327 96 + 27905053 C-U---GAUUUCCAGCCCGAG----AAUCUAAGGCUGGCGUUGCGCUUUACUCAGGGUCCGCUCGGCAACUGA-GCA-UUUAAAGUAAGCGCCGGCUAAGUUUAGU .-.---((((..(.....)..----))))...((((((((((..(((((((((((...((....))...))))-)..-..)))))).))))))))))......... ( -33.20) >DroVir_CAF1 40208 97 + 1 U-U---AAACGCCGGCUUUGG----GGCCUCAGGCUUGAGUUGCGUUUUACUCAGGGGUUGCGCGUCAACUGAAGCA-UUUGAAGUAGGCGCCGGCUAGGUUUAGU (-(---((((((((((.((..----.((.(((.((((.((((((((...(((....)))...))).))))).)))).-..))).)).)).))))))...)))))). ( -36.00) >DroPse_CAF1 26617 101 + 1 U-U---AUAUUCCAGCUCUGGGAGUAGCCUAAGGCUUUCGUUGCGUUUUGUUCAGGGGCUGGUCGGCAACUGA-ACAAUUUCAAGUAAGCGCCGGCUAAGUUUAGU .-.---...(((((....))))).(((((...(((.((..((.....((((((((..(((....)))..))))-)))).....))..)).))))))))........ ( -32.00) >DroGri_CAF1 42294 98 + 1 UUU---UGAAGCUGGCUUUGA----UGCCUUAGGCUUGAGUUGCGUUUUACUCAGGGGUUGCGCGUCUACUGAAACA-UUUGAAGUAGGCGCCGGCUAAGUUUAGU ...---..((((((((.....----.))).....(((((((........)))))))(((((.(((((((((......-.....)))))))))))))).)))))... ( -33.80) >DroAna_CAF1 25129 97 + 1 A-UAAGAAUUUCCAGCCUG------AAUUUAAGGCUGGCGUUGCGCUUUACUCAGGGUCCGCUCGGAAACUGA-GCA-UUUAAAGUAAGCGCCGGCUAAGUUUAGU .-..............(((------(((((..((((((((((..(((((((((((..(((....)))..))))-)..-..)))))).)))))))))))))))))). ( -39.90) >DroPer_CAF1 26629 101 + 1 G-U---AUAUUCCAGCUCUGGGAGUAGCCUAAGGCUUUCGUUGCGUUUUGUUCAGGGGCUGGUCGGCAACUGA-ACAAUUUCAAGUAAGCGCCGGCUAAGUUUAGU .-.---...(((((....))))).(((((...(((.((..((.....((((((((..(((....)))..))))-)))).....))..)).))))))))........ ( -32.00) >consensus U_U___AAAUUCCAGCUCUGG____AGCCUAAGGCUUGCGUUGCGUUUUACUCAGGGGCUGCUCGGCAACUGA_ACA_UUUAAAGUAAGCGCCGGCUAAGUUUAGU .............((((........((((...))))......((((((...((((..(((....)))..)))).((........)))))))).))))......... (-16.62 = -15.57 + -1.05)
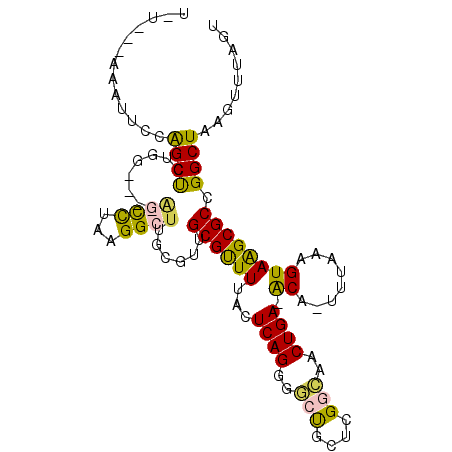
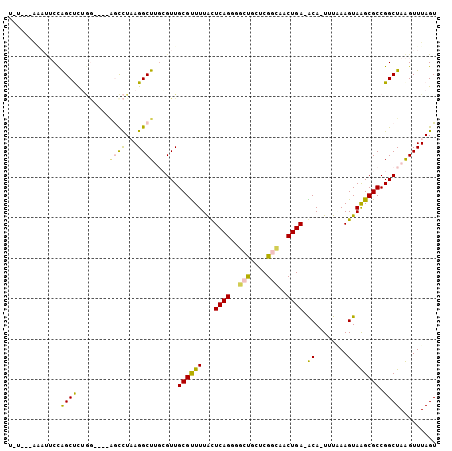
| Location | 3,341,327 – 3,341,423 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.05 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3341327 96 - 27905053 ACUAAACUUAGCCGGCGCUUACUUUAAA-UGC-UCAGUUGCCGAGCGGACCCUGAGUAAAGCGCAACGCCAGCCUUAGAUU----CUCGGGCUGGAAAUC---A-G ..............((((((........-(((-((((...((....))...)))))))))))))....(((((((.((...----)).))))))).....---.-. ( -31.50) >DroVir_CAF1 40208 97 - 1 ACUAAACCUAGCCGGCGCCUACUUCAAA-UGCUUCAGUUGACGCGCAACCCCUGAGUAAAACGCAACUCAAGCCUGAGGCC----CCAAAGCCGGCGUUU---A-A ..(((((...((((((.....(((((..-.((((.(((((.((....((......))....))))))).)))).)))))..----.....))))))))))---)-. ( -28.30) >DroPse_CAF1 26617 101 - 1 ACUAAACUUAGCCGGCGCUUACUUGAAAUUGU-UCAGUUGCCGACCAGCCCCUGAACAAAACGCAACGAAAGCCUUAGGCUACUCCCAGAGCUGGAAUAU---A-A ........(((((...((((.((((...((((-((((..((......))..))))))))....))).).))))....)))))...(((....))).....---.-. ( -25.40) >DroGri_CAF1 42294 98 - 1 ACUAAACUUAGCCGGCGCCUACUUCAAA-UGUUUCAGUAGACGCGCAACCCCUGAGUAAAACGCAACUCAAGCCUAAGGCA----UCAAAGCCAGCUUCA---AAA .....((((((...(((((((((.....-......)))))..)))).....))))))............((((....(((.----.....))).))))..---... ( -20.40) >DroAna_CAF1 25129 97 - 1 ACUAAACUUAGCCGGCGCUUACUUUAAA-UGC-UCAGUUUCCGAGCGGACCCUGAGUAAAGCGCAACGCCAGCCUUAAAUU------CAGGCUGGAAAUUCUUA-U ..............((((((........-(((-((((..(((....)))..)))))))))))))....(((((((......------.))))))).........-. ( -33.70) >DroPer_CAF1 26629 101 - 1 ACUAAACUUAGCCGGCGCUUACUUGAAAUUGU-UCAGUUGCCGACCAGCCCCUGAACAAAACGCAACGAAAGCCUUAGGCUACUCCCAGAGCUGGAAUAU---A-C ........(((((...((((.((((...((((-((((..((......))..))))))))....))).).))))....)))))...(((....))).....---.-. ( -25.40) >consensus ACUAAACUUAGCCGGCGCUUACUUCAAA_UGC_UCAGUUGCCGAGCAGCCCCUGAGUAAAACGCAACGCAAGCCUUAGGCU____CCAAAGCUGGAAUUU___A_A ...........(((((.............(((.((((..((......))..)))))))....((.......)).................)))))........... (-12.52 = -12.05 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:57 2006