
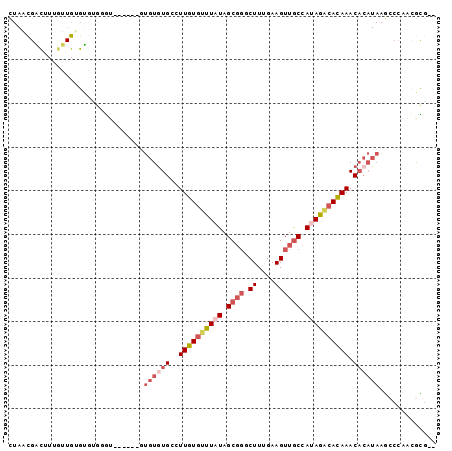
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,334,563 – 3,334,655 |
| Length | 92 |
| Max. P | 0.681765 |

| Location | 3,334,563 – 3,334,655 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -11.79 |
| Energy contribution | -13.57 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3334563 92 + 27905053 CUAACGACUUUGUUGUGUAUGCGU------GUGUGUGCCUUGUGUUUAUAGCGGGCUUUGAAGUUGCCAUAGACACAAACACAUAAGCCCAACGCGUU ...(((((...)))))..((((((------.((((((..((((((((((.((((.((....)))))).)))))))))).))))))......)))))). ( -28.40) >DroSec_CAF1 28659 98 + 1 CUAACGACUUUGUUGAGUGUGGGUGUGUGUGUGUGUGCCUUGUGUUUAUAGCGGGCUUUGAAGUUGCCAUAGACACAAACACAUAAGCCCAAAGCGUU ..((((((((....)))).(((((...(((((((.....((((((((((.((((.((....)))))).))))))))))))))))).)))))...)))) ( -32.60) >DroSim_CAF1 22558 94 + 1 CUAACGACUUUAUUGUGUGUGGGUGUGA--GUGUGUGCCUUGUGUUUAUAGCGGGCUUUGAAGUUGCCAUAGACACAAACACAUAAGCCCAACGCG-- ..............((((.(((((....--.((((((..((((((((((.((((.((....)))))).)))))))))).)))))).))))))))).-- ( -32.70) >DroWil_CAF1 27443 80 + 1 ---UUGGC-UGGCUGGGUACGAAC------CGGUGUGCAUUGUUUUUUUAGCCUCCUUUGAAGUUACCACAAGCAUAAAC--------CAAAUAUGGU ---((((.-..(((((.......)------))))((((.((((.....((((.((....)).))))..))))))))...)--------)))....... ( -15.90) >DroYak_CAF1 23159 90 + 1 CUAACGACUUUGUUGUGUGGGUGGG------UGUAUGUCUUGUGUUUAUAGCGGUCUUUGAAGUUGCCAUAGUCACAAACACAUAAGCCCCGCGUG-- ((((((((...))))).)))(((((------..(((((.(((((.((((.((((.((....)))))).)))).)))))..)))))...)))))...-- ( -25.10) >DroAna_CAF1 19150 90 + 1 UUAACGUUUUGACUGAGAGGUUAU------UUGUGUGAAUUGUGUUUUUAGCGGCCUUUGAAGUUGCCAUAGACACAAACACAUAAACACAACAUC-- .....(((.(((((....)))))(------(((((((..((((((((...(((((.......)))))...)))))))).))))))))...)))...-- ( -25.00) >consensus CUAACGACUUUGUUGUGUGUGGGU______GUGUGUGCCUUGUGUUUAUAGCGGGCUUUGAAGUUGCCAUAGACACAAACACAUAAGCCCAACGCG__ ...............................((((((..((((((((((.((((.((....)))))).)))))))))).))))))............. (-11.79 = -13.57 + 1.78)


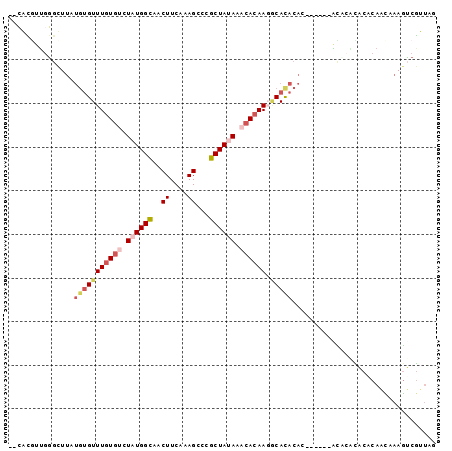
| Location | 3,334,563 – 3,334,655 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -11.54 |
| Energy contribution | -12.85 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3334563 92 - 27905053 AACGCGUUGGGCUUAUGUGUUUGUGUCUAUGGCAACUUCAAAGCCCGCUAUAAACACAAGGCACACAC------ACGCAUACACAACAAAGUCGUUAG ...((((((.((((.((((((((((.(...(((.........))).).))))))))))))))...)).------)))).................... ( -23.70) >DroSec_CAF1 28659 98 - 1 AACGCUUUGGGCUUAUGUGUUUGUGUCUAUGGCAACUUCAAAGCCCGCUAUAAACACAAGGCACACACACACACACCCACACUCAACAAAGUCGUUAG (((((((((((....(((((.((((((((((((.............))))).......))))))).)))))....)))).........))).)))).. ( -24.33) >DroSim_CAF1 22558 94 - 1 --CGCGUUGGGCUUAUGUGUUUGUGUCUAUGGCAACUUCAAAGCCCGCUAUAAACACAAGGCACACAC--UCACACCCACACACAAUAAAGUCGUUAG --.(.((((((....(((((((((((.((((((.............)))))).)))))).)))))...--.....)))).)).).............. ( -22.12) >DroWil_CAF1 27443 80 - 1 ACCAUAUUUG--------GUUUAUGCUUGUGGUAACUUCAAAGGAGGCUAAAAAAACAAUGCACACCG------GUUCGUACCCAGCCA-GCCAA--- .......(((--------(((...(((.(.((((....(...((.(((............)).).)).------)....)))))))).)-)))))--- ( -15.20) >DroYak_CAF1 23159 90 - 1 --CACGCGGGGCUUAUGUGUUUGUGACUAUGGCAACUUCAAAGACCGCUAUAAACACAAGACAUACA------CCCACCCACACAACAAAGUCGUUAG --......(((..(((((.((((((..((((((.............))))))..)))))))))))..------)))...................... ( -20.42) >DroAna_CAF1 19150 90 - 1 --GAUGUUGUGUUUAUGUGUUUGUGUCUAUGGCAACUUCAAAGGCCGCUAAAAACACAAUUCACACAA------AUAACCUCUCAGUCAAAACGUUAA --((((((.(((((.((((.((((((.(.((((..((.....))..)))).).))))))..)))).))------))).............)))))).. ( -17.44) >consensus __CACGUUGGGCUUAUGUGUUUGUGUCUAUGGCAACUUCAAAGCCCGCUAUAAACACAAGGCACACAC______ACACACACACAACAAAGUCGUUAG ...............(((((((((((.((((((..((....))...)))))).)))))).)))))................................. (-11.54 = -12.85 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:53 2006