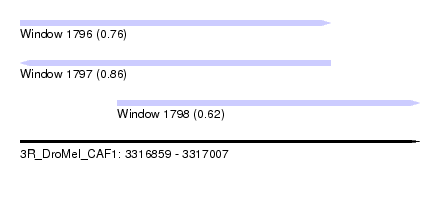
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,316,859 – 3,317,007 |
| Length | 148 |
| Max. P | 0.860960 |

| Location | 3,316,859 – 3,316,974 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.22 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3316859 115 + 27905053 AAAAUAAAAAGUGUU----UCAUCACAUUUUUUGUAAUAUUUAUAAAUCAAGCACUUCUGCAACACUAUUUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUU ...(((((((((((.----.....)))))))))))...............(((((((((((..((((........))))..((.((((((((....)))))))))).))))))).)))) ( -33.70) >DroSec_CAF1 10961 114 + 1 UAAAAAAAAU-UUUU----UCAUCACAUUUUUUGUAAUAUCUAUAAAUCAAGCACUUCUUCAAAACUAUUUUCCCAAUGCACACGCAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUU ..........-....----...........((((((.....))))))...(((((((((...................((.((.((((((((....)))))))))).))))))).)))) ( -19.40) >DroSim_CAF1 4271 115 + 1 UAAAUAAAAUAUGUU----UCAUCAAAUUUUUUGUUAUAUUUAUAAAUCAAGCACUUCUUUAAAACUAUUUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUU ((((((.((((.(((----(....))))....)))).)))))).......(((((((((.....(((........)))((.((.((((((((....)))))))))).))))))).)))) ( -20.90) >DroYak_CAF1 4432 105 + 1 -AAAUAGAAGUCACUGGACUGCUU-AAGUUUUUUUAAUAUUU------------CUUCUUCACAAAUAUUUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGUUGGGCAGAAGCUAUU -.(((((...((((((((((....-.)))......(((((((------------.........)))))))...)))))((.(...(((((((....)))))))...))).))..))))) ( -19.90) >consensus UAAAUAAAAU_UGUU____UCAUCAAAUUUUUUGUAAUAUUUAUAAAUCAAGCACUUCUUCAAAACUAUUUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUU ....................................................................((((((((((....))((((((((....)))))))))))).))))...... (-14.35 = -14.22 + -0.13)

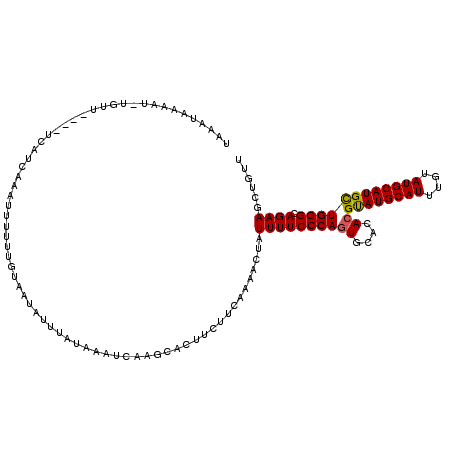
| Location | 3,316,859 – 3,316,974 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3316859 115 - 27905053 AACAGCUUCUGCCCAGCAUGCAUACAAAUGCAUACGUGUGCACUGGGAAAAUAGUGUUGCAGAAGUGCUUGAUUUAUAAAUAUUACAAAAAAUGUGAUGA----AACACUUUUUAUUUU ...........(((((..(((((((..........)))))))))))).(((.(((((((((....)))............(((((((.....))))))).----)))))).)))..... ( -27.50) >DroSec_CAF1 10961 114 - 1 AACAGCUUCUGCCCAGCAUGCAUACAAAUGCAUGCGUGUGCAUUGGGAAAAUAGUUUUGAAGAAGUGCUUGAUUUAUAGAUAUUACAAAAAAUGUGAUGA----AAAA-AUUUUUUUUA ....((((((((.((((((((((....)))))))).)).)).(..(((......)))..)))))))..............(((((((.....))))))).----....-.......... ( -25.80) >DroSim_CAF1 4271 115 - 1 AACAGCUUCUGCCCAGCAUGCAUACAAAUGCAUACGUGUGCACUGGGAAAAUAGUUUUAAAGAAGUGCUUGAUUUAUAAAUAUAACAAAAAAUUUGAUGA----AACAUAUUUUAUUUA ...........(((((..(((((((..........))))))))))))((((((((((((..(((....(((..((((....)))))))....)))..)))----))).))))))..... ( -22.10) >DroYak_CAF1 4432 105 - 1 AAUAGCUUCUGCCCAACAUGCAUACAAAUGCAUACGUGUGCACUGGGAAAAUAUUUGUGAAGAAG------------AAAUAUUAAAAAAACUU-AAGCAGUCCAGUGACUUCUAUUU- ....((((..(((((...(((((((..........))))))).))))..(((((((.(.....).------------))))))).......)..-))))((((....)))).......- ( -21.50) >consensus AACAGCUUCUGCCCAGCAUGCAUACAAAUGCAUACGUGUGCACUGGGAAAAUAGUUUUGAAGAAGUGCUUGAUUUAUAAAUAUUACAAAAAAUGUGAUGA____AACA_AUUUUAUUUA ....((((((.(((((..(((((((..........)))))))))))).............))))))..................................................... (-18.17 = -18.54 + 0.37)

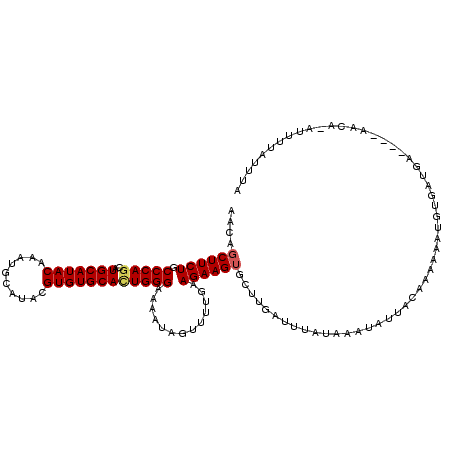
| Location | 3,316,895 – 3,317,007 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -16.16 |
| Energy contribution | -15.50 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3316895 112 + 27905053 UUAUAAAUCAAGCACUUCUGCAACACUAU-UUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUUGCAAAACCA-------AGACGCGAGACCUCAAAACACAGA ..........(((((((((((..((((..-......))))..((.((((((((....)))))))))).))))))).)))).........-------...(....)............... ( -27.60) >DroSec_CAF1 10996 112 + 1 CUAUAAAUCAAGCACUUCUUCAAAACUAU-UUUCCCAAUGCACACGCAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUUGCAAAACCA-------AGACGCGAGACCUCAAAACACAGA ...........((((..(((((((....)-))......(((.((.((((((((....)))))))))).)))))))..).))).......-------...(....)............... ( -21.90) >DroSim_CAF1 4307 112 + 1 UUAUAAAUCAAGCACUUCUUUAAAACUAU-UUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUGUUGCAAAACCA-------AGACGCGAGACCUCAAAACACAGA .......((..((...............(-(((((((((....))((((((((....)))))))))))).))))((....)).......-------....))..)).............. ( -21.10) >DroEre_CAF1 4361 100 + 1 UU------------CUUGCUCAAAAAUAU-UUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUAUUGCAAAACCA-------AGACGCGAGACCUCAAAACAGAGA .(------------(((((((.......(-(((((((((....))((((((((....)))))))))))).))))((....)).......-------.)).)))))).(((......))). ( -28.70) >DroYak_CAF1 4470 100 + 1 UU------------CUUCUUCACAAAUAU-UUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGUUGGGCAGAAGCUAUUGCAAAACCA-------AGGCGCGAGACCUCAAAACAGGGA ..------------...............-..((((.((((....((((((....)))))).(((..(((....)))...)))......-------..))))((....))......)))) ( -23.30) >DroAna_CAF1 3800 99 + 1 ---------------------AGAUCGAUUUCUCGAAGUGCAUAGACAUGCAUUUGUAUGCAUGUGAGGCAGAAGGUAUUGCAACACCACGACGCGAGACGCGAGACCUCAAAACACAGA ---------------------...((((....)))).(((.....((((((((....))))))))(((((....(((........)))....((((...)))).).))))....)))... ( -27.50) >consensus UU____________CUUCUUCAAAACUAU_UUUCCCAGUGCACACGUAUGCAUUUGUAUGCAUGCUGGGCAGAAGCUAUUGCAAAACCA_______AGACGCGAGACCUCAAAACACAGA ..............................((((....(((.((.((((((((....)))))))))).)))...((....))....................)))).............. (-16.16 = -15.50 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:51 2006