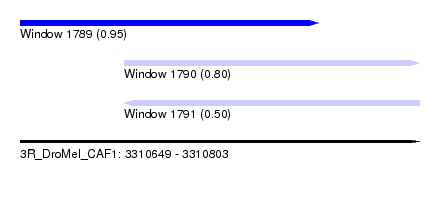
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,310,649 – 3,310,803 |
| Length | 154 |
| Max. P | 0.952073 |

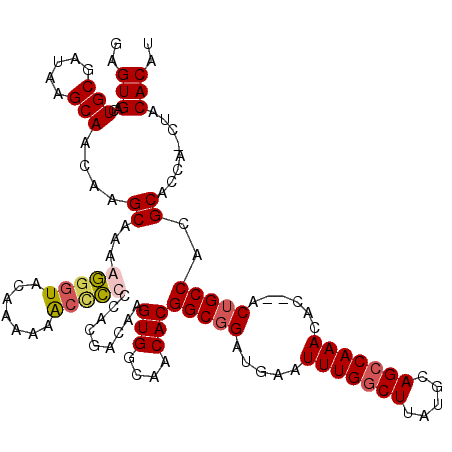
| Location | 3,310,649 – 3,310,764 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -27.62 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
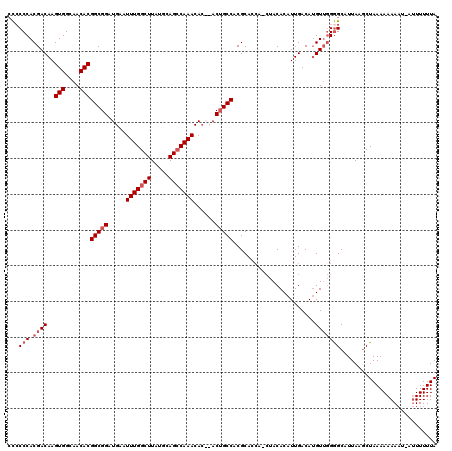
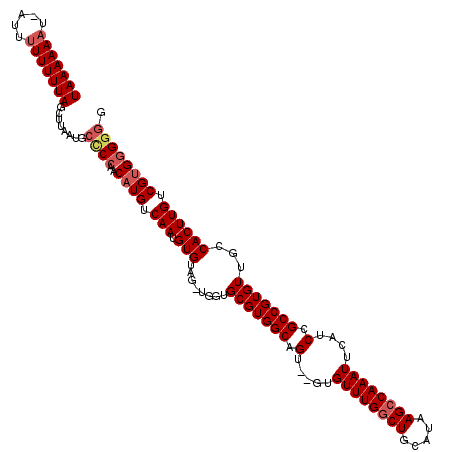
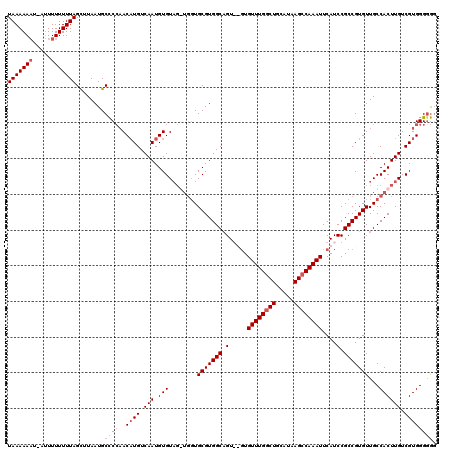
>3R_DroMel_CAF1 3310649 115 + 27905053 GAGUGACUGCGAUGAGCAACAAGCAAAAGGGUACAAAAAGCUCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACAC--ACGGCCACGCAGCA---ACACAU ..(((.(((((.((.((.....))....(((..(.....)..))).....)).(((....)))(((.(.((..(((((((.....)))))))..)--)).))).)))))..---.))).. ( -35.70) >DroSec_CAF1 14822 119 + 1 GAGUGACUGCGAUAAGCAACAAGCGAAGUGGUA-AAAAAACCGCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACACACACUGCCACGCACCACCUACACAU ..(((..(((.....)))....(((..(((((.-.....))))).........(((....)))(((((.((..(((((((.....)))))))..))..))))).)))........))).. ( -37.70) >DroSim_CAF1 19065 120 + 1 GAGUGACUGCGAUAAGCAACAAGCGAAGGGGUAUAAAAAACCCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACACACACUGCCACGCACCACCUACACAU ..(((..(((.....)))....(((..(((((.......))))).........(((....)))(((((.((..(((((((.....)))))))..))..))))).)))........))).. ( -39.90) >DroYak_CAF1 25084 113 + 1 GAGUGACUGCGAUAAGCAACAAGCAAAAGAGUGCAAAAAG-CCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGGCAAACA---ACUGCCACGCAC---CUACACAU ..(((..(((.....))).........((.((((......-............(((....)))(((((.((..((((.((.....)).))))))---.)))))..))))---)).))).. ( -30.90) >consensus GAGUGACUGCGAUAAGCAACAAGCAAAAGGGUACAAAAAACCCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACAC__ACUGCCACGCACCA_CUACACAU ..(((..(((.....)))....((...(((((.......))))).........(((....)))(((((.....(((((((.....)))))))......)))))..))........))).. (-27.62 = -28.38 + 0.75)

| Location | 3,310,689 – 3,310,803 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.53 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.82 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3310689 114 + 27905053 CUCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACAC--ACGGCCACGCAGCA---ACACAUUGACAUGUUCGAGCAUUAAGCUAAAAAAAAC-AUUUUUUA .......(((((.(((....)))(((.(.((..(((((((.....)))))))..)--)).)))........---...........)).)))(((.....))).........-........ ( -26.30) >DroSec_CAF1 14861 120 + 1 CCGCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACACACACUGCCACGCACCACCUACACAUUGACAGGUUGGGGCAUUAAGCUAAAAAAAAUGAUUUUUUA ..((((.((((..(((....)))(((((.((..(((((((.....)))))))..))..))))).......................)))))))).......(((((((.....))))))) ( -35.80) >DroSim_CAF1 19105 120 + 1 CCCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACACACACUGCCACGCACCACCUACACAUUGACAUGUUGGGGCAUUAAGCUAAAAAAAAUGAUUUUUUA ...(((.(((((.(((....)))(((((.((..(((((((.....)))))))..))..)))))......................))))))))........(((((((.....))))))) ( -32.10) >DroYak_CAF1 25124 110 + 1 -CCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGGCAAACA---ACUGCCACGCAC---CUACACAUUGACAUGUGGGGGCAUUAAACUAAAAAAAC--A-UUUUUA -((((((((....(((....)))(((((.((..((((.((.....)).))))))---.)))))......---.............)))))))).................--.-...... ( -33.30) >consensus CCCCCCACGACAAGUGGCAACACGGCGGAUGAAUUUGGCUUAUGCAGCCAAACAC__ACUGCCACGCACCA_CUACACAUUGACAUGUUGGGGCAUUAAGCUAAAAAAAAU_AUUUUUUA ...(((.((((..(((....)))(((((.....(((((((.....)))))))......))))).......................)))))))........................... (-22.58 = -23.82 + 1.25)

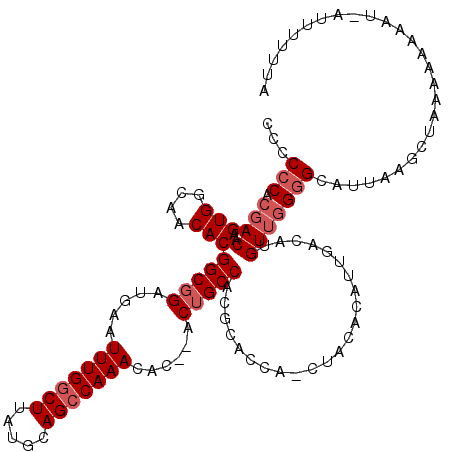
| Location | 3,310,689 – 3,310,803 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.53 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -25.17 |
| Energy contribution | -26.55 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3310689 114 - 27905053 UAAAAAAU-GUUUUUUUUAGCUUAAUGCUCGAACAUGUCAAUGUGU---UGCUGCGUGGCCGU--GUGUUUGGCUGCAUAAGCCAAAUUCAUCCGCCGUGUUGCCACUUGUCGUGGGGAG (((((((.-...)))))))........(((...((((.(((.(((.---.((.(((((((.((--(.((((((((.....)))))))).)))..))))))).)))))))).))))..))) ( -35.50) >DroSec_CAF1 14861 120 - 1 UAAAAAAUCAUUUUUUUUAGCUUAAUGCCCCAACCUGUCAAUGUGUAGGUGGUGCGUGGCAGUGUGUGUUUGGCUGCAUAAGCCAAAUUCAUCCGCCGUGUUGCCACUUGUCGUGGGCGG (((((((.....)))))))(((((..((.(((.((((.(.....)))))))).))(((((((..((.((((((((.....))))))))........))..)))))))......))))).. ( -39.30) >DroSim_CAF1 19105 120 - 1 UAAAAAAUCAUUUUUUUUAGCUUAAUGCCCCAACAUGUCAAUGUGUAGGUGGUGCGUGGCAGUGUGUGUUUGGCUGCAUAAGCCAAAUUCAUCCGCCGUGUUGCCACUUGUCGUGGGGGG (((((((.....)))))))........(((((((((....))))((((((((((((((((.(..((.((((((((.....)))))))).))..))))))).))))))))).).))))).. ( -43.50) >DroYak_CAF1 25124 110 - 1 UAAAAA-U--GUUUUUUUAGUUUAAUGCCCCCACAUGUCAAUGUGUAG---GUGCGUGGCAGU---UGUUUGCCUGCAUAAGCCAAAUUCAUCCGCCGUGUUGCCACUUGUCGUGGGGG- ((((((-.--...))))))........((((((((((....((.((..---(((((.(((((.---...))))))))))..))))....))).....(((....))).....)))))))- ( -31.70) >consensus UAAAAAAU_AUUUUUUUUAGCUUAAUGCCCCAACAUGUCAAUGUGUAG_UGGUGCGUGGCAGU__GUGUUUGGCUGCAUAAGCCAAAUUCAUCCGCCGUGUUGCCACUUGUCGUGGGGGG (((((((.....)))))))........((((..((((.(((.(((........(((((((.(.....((((((((.....))))))))....).)))))))...)))))).)))))))). (-25.17 = -26.55 + 1.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:44 2006