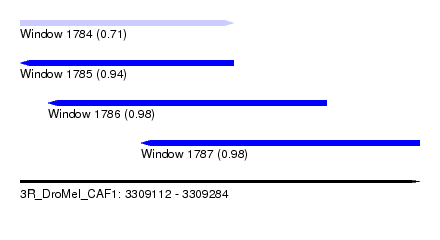
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,309,112 – 3,309,284 |
| Length | 172 |
| Max. P | 0.983097 |

| Location | 3,309,112 – 3,309,204 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.47 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3309112 92 + 27905053 AGUGCAGAAUCAAGGAUCUUGAGGCCAUGGAGCUGAAGCCUGGAGCUGGAGCACCAACGCCACAUAAAUUGCGGCGCCCAAAAUGCAUAAAC .(((((...(((((...)))))(((..(((.(((..(((.....)))..))).)))..(((.((.....)).)))))).....))))).... ( -29.70) >DroSec_CAF1 13323 79 + 1 AGUGCAGAAUCAAGGAUCUUGAGGCCAUGGA-------------GCUGGAGCACCAACGCCACAUAAAUUGCGGCGCCCAAAAUGCAUAAAC .(((((...(((((...)))))(((..(((.-------------((....)).)))..(((.((.....)).)))))).....))))).... ( -23.90) >DroSim_CAF1 17574 79 + 1 AGUGCAGAAUCAAGGAUCUUGAGGCCAUGGA-------------GCUGGAGCACCAACGCCACAUAAAUUGCGGCGCCCAAAAUGCAUAAAC .(((((...(((((...)))))(((..(((.-------------((....)).)))..(((.((.....)).)))))).....))))).... ( -23.90) >DroYak_CAF1 23602 85 + 1 AGUGCAGAACCAAGAAUCUUGAGGCCAUGGAGCC-------GGAGCCGGAGCACCAACGCCACAUAAAUUGCGACGCCCAAAAUGCAUAAAG .(((((....((((...)))).(((..(((..((-------(....)))....))).(((..........)))..))).....))))).... ( -19.30) >consensus AGUGCAGAAUCAAGGAUCUUGAGGCCAUGGA_____________GCUGGAGCACCAACGCCACAUAAAUUGCGGCGCCCAAAAUGCAUAAAC .((((((..(((((...)))))..)..(((..............((....)).....((((.((.....)).)))).)))...))))).... (-16.98 = -17.47 + 0.50)
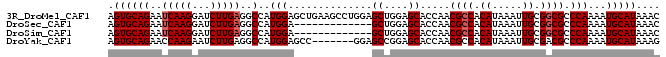
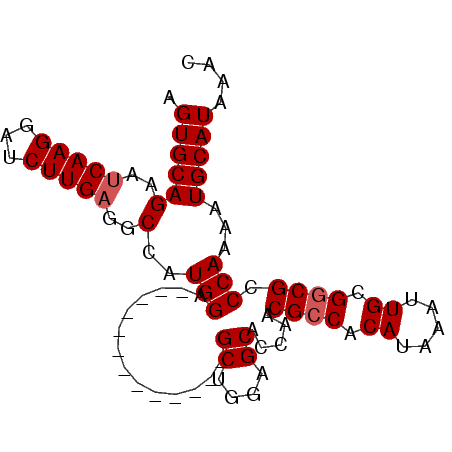
| Location | 3,309,112 – 3,309,204 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3309112 92 - 27905053 GUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGCUCCAGGCUUCAGCUCCAUGGCCUCAAGAUCCUUGAUUCUGCACU .....((((.....(((((((((.....))))))..(((.(((..(((.....)))..))).)))..)))(((((...)))))...)))).. ( -30.30) >DroSec_CAF1 13323 79 - 1 GUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGC-------------UCCAUGGCCUCAAGAUCCUUGAUUCUGCACU .....((((.....(((((((((.....))))))(((((....)))))-------------......)))(((((...)))))...)))).. ( -25.80) >DroSim_CAF1 17574 79 - 1 GUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGC-------------UCCAUGGCCUCAAGAUCCUUGAUUCUGCACU .....((((.....(((((((((.....))))))(((((....)))))-------------......)))(((((...)))))...)))).. ( -25.80) >DroYak_CAF1 23602 85 - 1 CUUUAUGCAUUUUGGGCGUCGCAAUUUAUGUGGCGUUGGUGCUCCGGCUCC-------GGCUCCAUGGCCUCAAGAUUCUUGGUUCUGCACU .....((((((((((((((((((.....))))))(((((.((....)).))-------)))......))).)))))..........)))).. ( -26.00) >consensus GUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGC_____________UCCAUGGCCUCAAGAUCCUUGAUUCUGCACU .....((((.....(((((((((.....))))))(((((....)))))...................)))(((((...)))))...)))).. (-24.66 = -24.10 + -0.56)

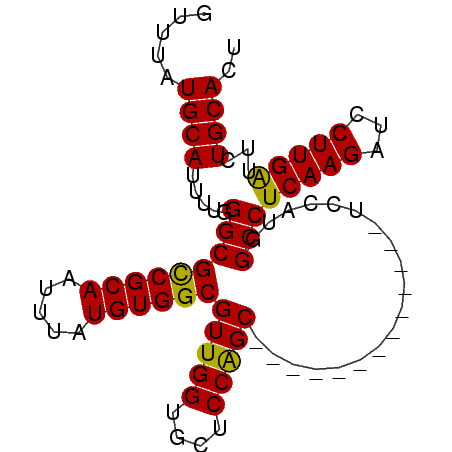
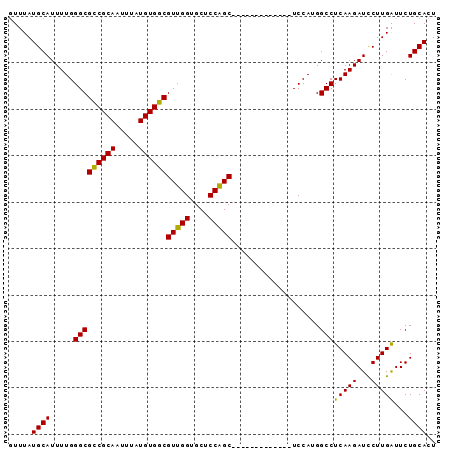
| Location | 3,309,124 – 3,309,244 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -39.79 |
| Energy contribution | -40.85 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3309124 120 - 27905053 GGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGCUCCAGGCUUCAGCUCCAUGGCCUCAAGAUCCU (((((.(((((.(((((((..((...((.(((.((((.........)))).))).))((((((.....))))))(((((....)))))..))...)))))))))))))))))........ ( -52.50) >DroSec_CAF1 13335 107 - 1 GGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGC-------------UCCAUGGCCUCAAGAUCCU (((((.(((((.(((((.........((.(((.((((.........)))).))).))((((((.....))))))..........))))-------------)))))))))))........ ( -44.20) >DroSim_CAF1 17586 107 - 1 GGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGC-------------UCCAUGGCCUCAAGAUCCU (((((.(((((.(((((.........((.(((.((((.........)))).))).))((((((.....))))))..........))))-------------)))))))))))........ ( -44.20) >DroYak_CAF1 23614 113 - 1 GGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGCUUUAUGCAUUUUGGGCGUCGCAAUUUAUGUGGCGUUGGUGCUCCGGCUCC-------GGCUCCAUGGCCUCAAGAUUCU (((((.(((((.(((((.........((.(((.((((.........)))).))).))((((((.....))))))(((((....)))))..)-------))))))))))))))........ ( -45.50) >consensus GGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGUGCUCCAGC_____________UCCAUGGCCUCAAGAUCCU (((((.(((((.((((..........((.(((.((((.........)))).))).))((((((.....))))))(((((....)))))..........))))))))))))))........ (-39.79 = -40.85 + 1.06)

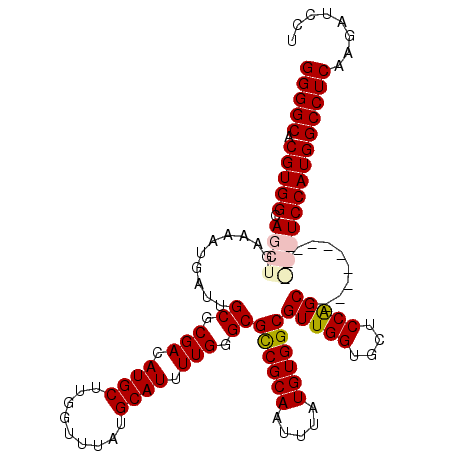
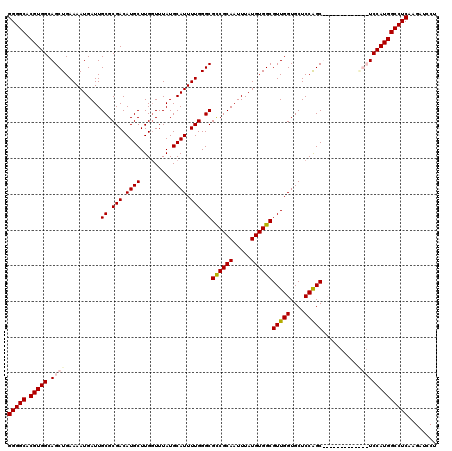
| Location | 3,309,164 – 3,309,284 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -46.05 |
| Consensus MFE | -43.28 |
| Energy contribution | -43.15 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3309164 120 - 27905053 UCCUUCCUUGCUUCCCCAUAAAGUGCCCGAAAUGUAUGCUGGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGU ...............(((......(((((((((((((((..((.((.((.((.((...........)))).)).))))..))..)))))))))))))((((((.....))))))..))). ( -44.30) >DroSec_CAF1 13362 117 - 1 UCCUUCCUUGC---CCCAUAAAGUGCCCGAAAUGUAUGCCGGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGU .....((.(((---(.(((((((((((((((((((((((((((.((.((.((.((...........)))).)).))))))))..))))))))))))))).....)))))).))))..)). ( -48.20) >DroSim_CAF1 17613 117 - 1 UCCUUCCUUGC---CCCAUAAAGUGCCCGAAAUGUAUGCCGGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGU .....((.(((---(.(((((((((((((((((((((((((((.((.((.((.((...........)))).)).))))))))..))))))))))))))).....)))))).))))..)). ( -48.20) >DroYak_CAF1 23647 117 - 1 UCCUCUCUCGC---CCCAUAAAGUGCCCGAAAUGUAUGCCGGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGCUUUAUGCAUUUUGGGCGUCGCAAUUUAUGUGGCGUUGGU ....((..(((---(.((((((.(((((((((((((((..((((((.((.((.((...........)))).)).))))).)..)))))))))))))))......)))))).))))..)). ( -43.50) >consensus UCCUUCCUUGC___CCCAUAAAGUGCCCGAAAUGUAUGCCGGGGCACGUGGCAGCUGAAAAUGAUUGCGCGACAUGCUUGGUUUAUGCAUUUUGGGCGCCGCAAUUUAUGUGGCGUUGGU ...............(((......(((((((((((((((((((.((.((.((.((...........)))).)).))))))))..)))))))))))))((((((.....))))))..))). (-43.28 = -43.15 + -0.13)

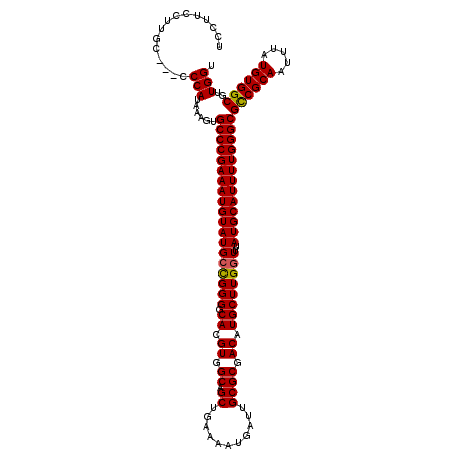
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:40 2006