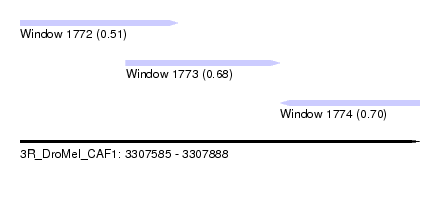
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,307,585 – 3,307,888 |
| Length | 303 |
| Max. P | 0.695670 |

| Location | 3,307,585 – 3,307,705 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3307585 120 + 27905053 GCUAUUUGUAAAUUUCGCGGCUGCAAUUAUGUUUGCUGUGUCGUGUUGCCAACAACCCCUCCCACCGCCACCGCCUGCCGACCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUC ((((..(((........((((.((((......)))).(((..(.((((....)))).)....)))...........))))........(((((....))))))))..))))......... ( -25.90) >DroSec_CAF1 11706 113 + 1 GCUAUUUGUAAAUUUCGCGGCUGCAAUUAUGUUUGCUGUGUCGUGUUGCCAACAACCCCUCCCACCAACACCGC-------CCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUC ((((..(((......((((((.(((....)))..))))))..((((((.................)))))).((-------(((....(....)...))))))))..))))......... ( -27.93) >DroSim_CAF1 15891 113 + 1 GCUAUUUGUAAAUUUCGCGGCUGCAAUUAUGUUUGCUGUGUCGUGUUGCCAACAACCCCUCCCACCAACACCGC-------CCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUC ((((..(((......((((((.(((....)))..))))))..((((((.................)))))).((-------(((....(....)...))))))))..))))......... ( -27.93) >consensus GCUAUUUGUAAAUUUCGCGGCUGCAAUUAUGUUUGCUGUGUCGUGUUGCCAACAACCCCUCCCACCAACACCGC_______CCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUC ((((..(((......((((((.((((......))))...))))))...........................................(((((....))))))))..))))......... (-25.10 = -25.10 + -0.00)

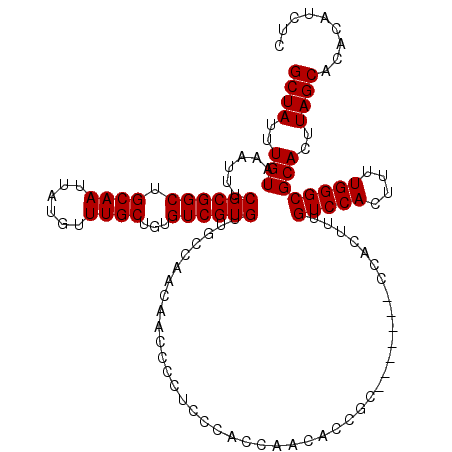
| Location | 3,307,665 – 3,307,782 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -20.29 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3307665 117 + 27905053 ACCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUCUGUCCAUCUCGCUCUGGCUUAACUUCCGGUUCCCUCGCAUUUU--UUUCUGGGCAUGUGGCUUUGCUGGUUAUUAUUAG ((((....(((((....)))))(((...(((.((((....((((((....((....))........(((.....)))......--....))))))))))))).)))))))......... ( -26.30) >DroSim_CAF1 15965 118 + 1 -CCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUCUGGCCAUCUCGCUCUGGCUUAACUUCCGGUUCCGUCACAUUUUUUUUUCUGGCCAUGUGGCUUUGCUGGUUAUUAUUAG -(((....(((((....)))))(((...(((.((((....((((((....((....))........((....))...............))))))))))))).)))))).......... ( -29.10) >DroYak_CAF1 22086 94 + 1 ----------------------GCACUUAGCACACAUCUCUGGCCAUCUCCCCCUGGCCUAACUUCCGGUUUCCUCGCACUU---UUUCUGGCCAUGUGGCUUUGCUGGCUAUUAUUAG ----------------------(((...(((.(((((....(((((........)))))........((((...........---.....)))))))))))).)))............. ( -21.19) >consensus _CCACUUUGUCCACUUUUGGGCGCACUUAGCACACAUCUCUGGCCAUCUCGCUCUGGCUUAACUUCCGGUUCCCUCGCAUUUU__UUUCUGGCCAUGUGGCUUUGCUGGUUAUUAUUAG ........(((((....)))))(((...(((.((((....((((((.......((((........))))....................))))))))))))).)))............. (-20.29 = -20.26 + -0.03)

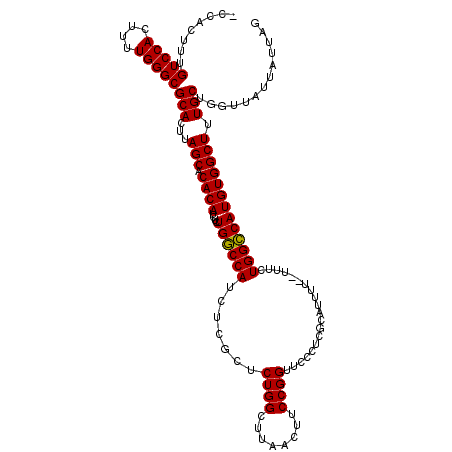
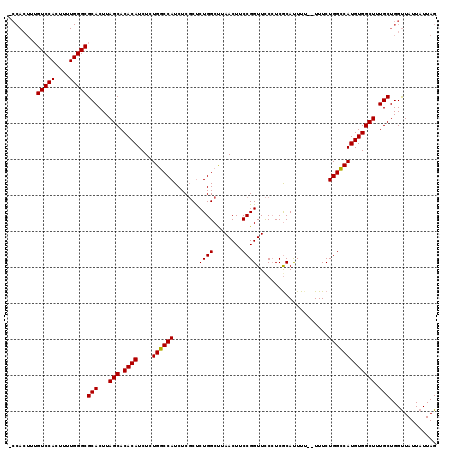
| Location | 3,307,782 – 3,307,888 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -16.82 |
| Energy contribution | -19.51 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
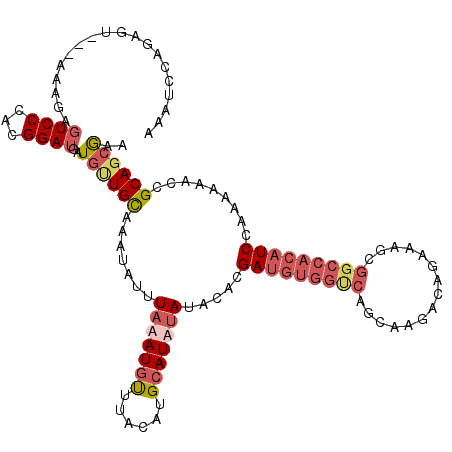
>3R_DroMel_CAF1 3307782 106 - 27905053 AAAUCCAGAGU---AAA--GUCCCACGGAUGCUGCUGCAAAUAUUUAAAUGUUUACAUGCAUAUAUACACGAUGUGGUCAGCAAGACAGAAG---------UCCACAAAACCGCAGCGAA ........(((---...--((((...)))))))(((((..........((((......))))..........(((((.(..(......)..)---------.))))).....)))))... ( -20.80) >DroSec_CAF1 11899 117 - 1 AAAUCCCGAGU---AAAGAGUCCCACGGAUGAUAUUGCAAAUAUUUAAAUGUUUACAUGCAUAUAUACACGAUGUGGUCAGCAAGACAGAAAGCGGCCACAUCCAAAAAACCGCAGCGAA ......((...---.....((((...))))....((((..........((((......))))........(((((((((.((..........))))))))))).........)))))).. ( -23.10) >DroSim_CAF1 16083 116 - 1 AAAUC-CGAGU---AAAGAGUCCCACGGAUGAUGUUGCAAAUAUUUAUAUGUUUACAUGCAUAUAUACACGAUGUGGUCAGCAAGACAGAAAGCGGCCACAUCCAAAAAACCGCAGCGAA ..(((-((...---...........)))))..((((((.......(((((((......))))))).....(((((((((.((..........))))))))))).........)))))).. ( -32.34) >DroYak_CAF1 22180 120 - 1 AAUCCCAGACUCACAAAAAUUCCCACGGACGAUGGUGUAAAUAUUUUUAUGCUUACAUGGAUAUAUACACGAUGUGGCCAGCAGGACAAAAACCGGCCACAUCCAAAAAACCGCAGCAAA .........................(((......(((((.(((((..((((....))))))))).)))))(((((((((....((.......))))))))))).......)))....... ( -28.00) >consensus AAAUCCAGAGU___AAAGAGUCCCACGGAUGAUGUUGCAAAUAUUUAAAUGUUUACAUGCAUAUAUACACGAUGUGGUCAGCAAGACAGAAAGCGGCCACAUCCAAAAAACCGCAGCGAA ...................((((...))))..((((((.......(((((((......))))))).....(((((((((...............))))))))).........)))))).. (-16.82 = -19.51 + 2.69)

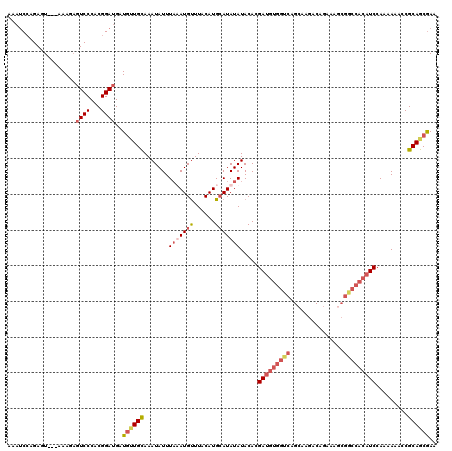
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:28 2006