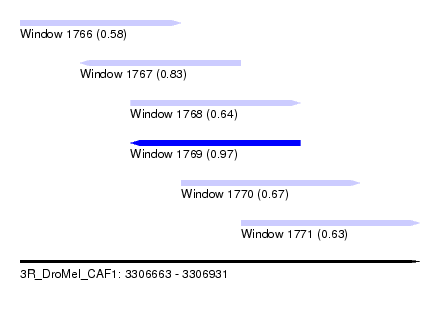
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,306,663 – 3,306,931 |
| Length | 268 |
| Max. P | 0.967454 |

| Location | 3,306,663 – 3,306,771 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3306663 108 + 27905053 AGAAGCCGGCAACGCCAAAUGUUAAGCAGGGUUUUGGGUUAGUGAGGCUCUGGCGGGGAUGGUGG------CCUCUGGGUUU------UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUU ...((((((((..((((..((..((((..(((((..((.....(((((((((.......))).))------)))).....))------..)))))..)))).))..))))..)))))))) ( -37.30) >DroSec_CAF1 10758 114 + 1 AGAAGCCGGCAACGCCAAAUGUUAAGCAGGGUUUUGGGUUAGUAGGGCUCUGGCGGGGAUGCUGGGUAGGUACUCUGGGUUU------UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUU ...((((((((...(((((.......((((((((((......)))))))))).(((((.((((.....)))))))))...))------)))((((((....)))))).....)))))))) ( -37.50) >DroSim_CAF1 14925 114 + 1 AGAAGCCGGCAACGCCAAAUGUUAAGCAGGGUUUUGGGUUAGUAGGGCUCUGGCGGGGAUGCUGGGUAGGUACUCUGGGUUU------UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUU ...((((((((...(((((.......((((((((((......)))))))))).(((((.((((.....)))))))))...))------)))((((((....)))))).....)))))))) ( -37.50) >DroYak_CAF1 20970 113 + 1 AGAAGCCGGCAACGCCUAAUGUUAAGCAGGGUUUUCGGUUAGUAAGGCUCUUUCGGGGAUGGGGGG-------UCUGGGUUUGGAGCUGGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUU ...((((((((..(((...((..((((..((((..(((((....((((.((..((....))..)))-------)))........)))))..))))..)))).))...)))..)))))))) ( -39.10) >consensus AGAAGCCGGCAACGCCAAAUGUUAAGCAGGGUUUUGGGUUAGUAAGGCUCUGGCGGGGAUGCUGGG_____ACUCUGGGUUU______UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUU ...((((((((..(((..........((((((((((......)))))))))).(((((..............)))))..............((((((....)))))))))..)))))))) (-29.50 = -29.82 + 0.31)

| Location | 3,306,703 – 3,306,811 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.831006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3306703 108 - 27905053 GUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA------AAACCCAGAGG------CCACCAUCCCCGCCAGAGCCUCACU .(((((((....(((.....)))...((((.((((((.....))))))))))..)))))))...........------.......((((------((.............).)))))... ( -29.62) >DroSec_CAF1 10798 114 - 1 GUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA------AAACCCAGAGUACCUACCCAGCAUCCCCGCCAGAGCCCUACU ((((((((....(((........((.((((.((((((.....)))))))))).))...........((((..------.......)))).........))).....))))))))...... ( -29.40) >DroSim_CAF1 14965 114 - 1 GUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA------AAACCCAGAGUACCUACCCAGCAUCCCCGCCAGAGCCCUACU ((((((((....(((........((.((((.((((((.....)))))))))).))...........((((..------.......)))).........))).....))))))))...... ( -29.40) >DroYak_CAF1 21010 113 - 1 CUUUUGGCAAGUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCCAGCUCCAAACCCAGA-------CCCCCCAUCCCCGAAAGAGCCUUACU .(((((((((((.((.....))))))((((.((((((.....))))))))))..)))))))....(((.....)))...........-------...........(....)......... ( -26.40) >consensus GUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA______AAACCCAGAGU_____CCCACCAUCCCCGCCAGAGCCCUACU .(((((((....(((.....)))...((((.((((((.....))))))))))..))))))).....((((........................................))))...... (-24.17 = -24.17 + 0.00)

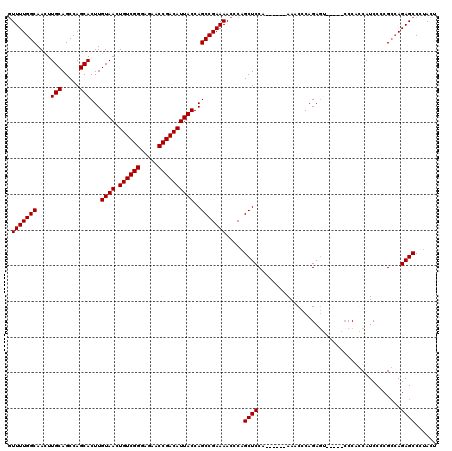
| Location | 3,306,737 – 3,306,851 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -38.35 |
| Energy contribution | -38.85 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3306737 114 + 27905053 UU------UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUUCUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUG ..------((..(((..((((..((((.((((((((((.....)))))).))))...(((((((((...((((......))))..))))))))))))))))).)))..)).......... ( -40.80) >DroSec_CAF1 10838 114 + 1 UU------UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUUCUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUG ..------((..(((..((((..((((.((((((((((.....)))))).))))...(((((((((...((((......))))..))))))))))))))))).)))..)).......... ( -40.80) >DroSim_CAF1 15005 114 + 1 UU------UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUUCUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUG ..------((..(((..((((..((((.((((((((((.....)))))).))))...(((((((((...((((......))))..))))))))))))))))).)))..)).......... ( -40.80) >DroYak_CAF1 21043 120 + 1 UUGGAGCUGGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUUCUCCCGACAGUUACAAGUGCUGGCUGCAACUUGCCAAAAGAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUG .....(((((....(((((((..((((.((((((((((.....)))))).))))...(((((((((...(((.......)))...)))))))))))))....))))).))....))))). ( -41.20) >consensus UU______UGGAGCUGGGUUUUCGGCUGGUAAUGUCGGUUCUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUG ..........(.(((..((((..((((.((((((((((.....)))))).))))...(((((((((...((((......))))..))))))))))))))))).))))............. (-38.35 = -38.85 + 0.50)

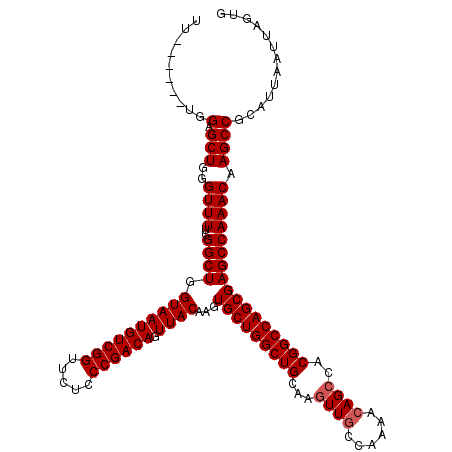
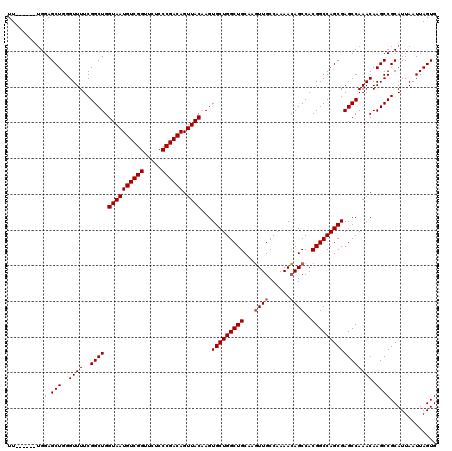
| Location | 3,306,737 – 3,306,851 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -38.83 |
| Energy contribution | -38.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3306737 114 - 27905053 CACUAAUUAAUGCGGCUUGUUUGGCUCGCUGGCCGUGGCUGUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA------AA ..........((.((((..(((((((.((((((.(..(.(((....)))..)..).))))))....((((.((((((.....)))))))))).))))))).....)))).))------.. ( -40.30) >DroSec_CAF1 10838 114 - 1 CACUAAUUAAUGCGGCUUGUUUGGCUCGCUGGCCGUGGCUGUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA------AA ..........((.((((..(((((((.((((((.(..(.(((....)))..)..).))))))....((((.((((((.....)))))))))).))))))).....)))).))------.. ( -40.30) >DroSim_CAF1 15005 114 - 1 CACUAAUUAAUGCGGCUUGUUUGGCUCGCUGGCCGUGGCUGUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA------AA ..........((.((((..(((((((.((((((.(..(.(((....)))..)..).))))))....((((.((((((.....)))))))))).))))))).....)))).))------.. ( -40.30) >DroYak_CAF1 21043 120 - 1 CACUAAUUAAUGCGGCUUGUUUGGCUCGCUGGCCGUGGCUCUUUUGGCAAGUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCCAGCUCCAA .............((....(((((((.((((((.(..(((.........)))..).))))))....((((.((((((.....)))))))))).)))))))...))(((.....))).... ( -42.10) >consensus CACUAAUUAAUGCGGCUUGUUUGGCUCGCUGGCCGUGGCUGUUUUGGCAACUUGCAGCCAGCACUUGUAACUGUCGGGAGAACCGACAUUACCAGCCGAAAACCCAGCUCCA______AA ...........((((....(((((((.((((((.((.(((.....))).....)).))))))....((((.((((((.....)))))))))).)))))))...)).))............ (-38.83 = -38.83 + -0.00)

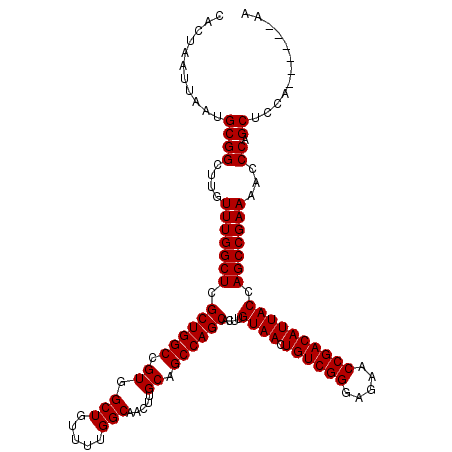
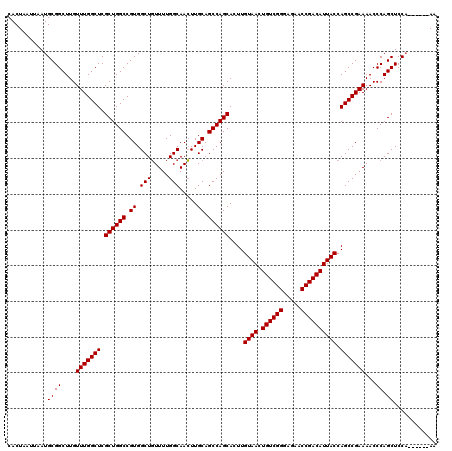
| Location | 3,306,771 – 3,306,891 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -35.80 |
| Energy contribution | -37.92 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3306771 120 + 27905053 CUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCC .(((.....(((.....(((((((((...((((......))))..))))))))).....)))..(((((........)))))...)))...((((.((..(((....)))..))..)))) ( -42.30) >DroSec_CAF1 10872 120 + 1 CUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCC .(((.....(((.....(((((((((...((((......))))..))))))))).....)))..(((((........)))))...)))...((((.((..(((....)))..))..)))) ( -42.30) >DroSim_CAF1 15039 120 + 1 CUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCC .(((.....(((.....(((((((((...((((......))))..))))))))).....)))..(((((........)))))...)))...((((.((..(((....)))..))..)))) ( -42.30) >DroYak_CAF1 21083 120 + 1 CUCCCGACAGUUACAAGUGCUGGCUGCAACUUGCCAAAAGAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACACGGACAUGGACGGAAUCC .(((.....(((.....(((((((((...(((.......)))...))))))))).....)))..(((((........)))))...(..((((..(........)..))))..)))).... ( -35.70) >consensus CUCCCGACAGUUACAAGUGCUGGCUGCAAGUUGCCAAAACAGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCC .(((.....(((.....(((((((((...((((......))))..))))))))).....)))..(((((........)))))...)))...((((.((..(((....)))..))..)))) (-35.80 = -37.92 + 2.13)

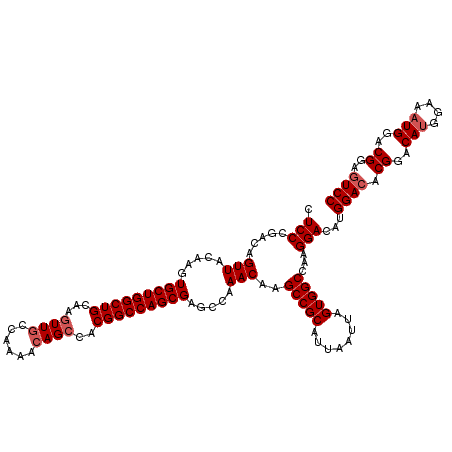
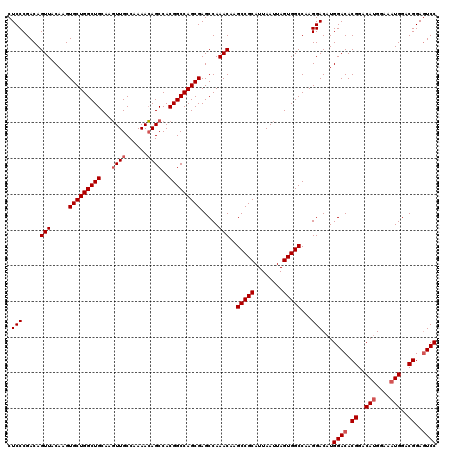
| Location | 3,306,811 – 3,306,931 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -36.49 |
| Energy contribution | -37.92 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3306811 120 + 27905053 AGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCCUGGCUCGUGCUCGCAGCUCCCGCAUUUUACAUUAACAAUU (((..((((..((((((((.....(((((........))))).........((((.((..(((....)))..))..)))))))))))))).))..)))...................... ( -41.40) >DroSec_CAF1 10912 120 + 1 AGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCCUGGCUCGUGCUCGCAGCUCCCGCAUUUUACAUUAACCAUU (((..((((..((((((((.....(((((........))))).........((((.((..(((....)))..))..)))))))))))))).))..)))...................... ( -41.40) >DroSim_CAF1 15079 120 + 1 AGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCCUGGCUCGUGCUCGCAGCUCCCGCAUUUUACAUUAACCAUU (((..((((..((((((((.....(((((........))))).........((((.((..(((....)))..))..)))))))))))))).))..)))...................... ( -41.40) >DroYak_CAF1 21123 119 + 1 AGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACACGGACAUGGACGGAAUCCUGGCUCGUGCUCGCAGUGCCCGC-UUUUACAUUAACAAUU ((((((.((..((((((((.....(((((........)))))...(..((((..(........)..))))..).......))))))))....)).)))...))-)............... ( -36.40) >consensus AGCCACGGCCAGCGAGCCAAACAAGCCGCAUUAAUUAGUGGCCAAGGACAUGGACACGGACAUGGAAAUGGACGGAGUCCUGGCUCGUGCUCGCAGCUCCCGCAUUUUACAUUAACAAUU .((..((((..((((((((.....(((((........))))).........((((.((..(((....)))..))..)))))))))))))).))..))....................... (-36.49 = -37.92 + 1.44)

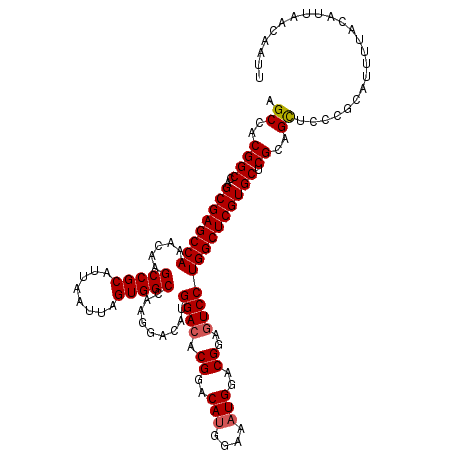
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:26 2006