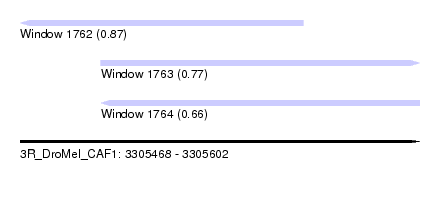
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,305,468 – 3,305,602 |
| Length | 134 |
| Max. P | 0.870866 |

| Location | 3,305,468 – 3,305,563 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -27.85 |
| Energy contribution | -27.73 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3305468 95 - 27905053 GCUGGUAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGAUG------------GGGGUGAACG-------------GUUGGCUGUAACUUUAUUUCAG .((((...((((((....))))))((((((.((.((((...))))(..((((((.......))------------))))..)..)-------------).))))))..........)))) ( -34.70) >DroSec_CAF1 9526 95 - 1 GCUGGCAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUACUAUUCUUGGUCCCAAGGACGAAG------------UGGGUGAACG-------------GUUGGCUGUAACUUUACUUCAG (((((((.....)))))))(((((((((((.((.(..((((...(((.((((...)))).)))------------..))))..))-------------).))))))).))))........ ( -36.70) >DroSim_CAF1 13135 95 - 1 GCUGGCAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGAAG------------UGGGUGAACG-------------GUUGGCUGUAACUUUAUUUCAG (((((((.....)))))))(((((((((((.((.....((((((((((....)))))).....------------)))).....)-------------).))))))).))))........ ( -33.70) >DroYak_CAF1 19735 120 - 1 GCUGGCAACACCUGUCAGCAGGUGCAGCAAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGCAGGUACGUUUGGUGGGGGAAAACGGUGCCAUCAAGCUGUUGGCUGUAACUUUAUUUCAG (((((((.....)))))))(((((((((.....(((((...)))))..((((((.(((((......)))))..))))))..((((((........)))))).))))).))))........ ( -40.20) >consensus GCUGGCAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGAAG____________GGGGUGAACG_____________GUUGGCUGUAACUUUAUUUCAG (((((((.....)))))))(((((((((((..(((....)))((((((....))))))..........................................))))))).))))........ (-27.85 = -27.73 + -0.13)

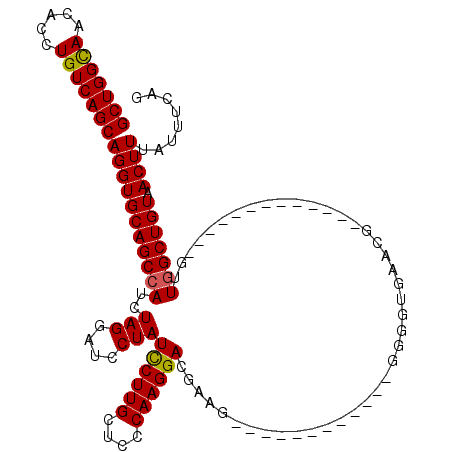
| Location | 3,305,495 – 3,305,602 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -29.23 |
| Energy contribution | -30.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3305495 107 + 27905053 ACCCC------------CAUCGUCCUUGGGAGCAAGGAUAGGAUCCUAGAUGGCUGCACCUGCUGACAGGUGUUACCAGCAUUUUGCAUAUGUCUUCC-UCCAUUCCUGGCAACUGAGCA ...((------------((.......)))).((.((((..(((....((((((..(((((((....)))))))..)).((.....))....))))...-)))..)))).))......... ( -34.70) >DroSec_CAF1 9553 107 + 1 ACCCA------------CUUCGUCCUUGGGACCAAGAAUAGUAUCCUAGAUGGCUGCACCUGCUGACAGGUGUUGCCAGCAUUUUGCAUAUGUCUUCC-UCCAUUUCUGUCAACUGAGCA .....------------(((.((((...)))).)))..((((....((((((((.(((((((....))))))).))))((.....))...........-......))))...)))).... ( -28.30) >DroSim_CAF1 13162 107 + 1 ACCCA------------CUUCGUCCUUGGGAGCAAGGAUAGGAUCCUAGAUGGCUGCACCUGCUGACAGGUGUUGCCAGCAUUUUGCAUAUGUCUUCC-UCCAUUUCUGUCAACUGAGCA .....------------.((((((((((....))))))).(((....(((((((.(((((((....))))))).))).((.....))....))))...-))).............))).. ( -34.70) >DroYak_CAF1 19775 120 + 1 UCCCCCACCAAACGUACCUGCGUCCUUGGGAGCAAGGAUAGGAUCCUAGAUUGCUGCACCUGCUGACAGGUGUUGCCAGCAUUUUGCAUAUGUCUUCCUUCCAUUUUUGGCAACUGAGCA ...((((....((((....))))...))))(((((...(((....)))..)))))(((((((....)))))(((((((((.....))..(((.........)))...)))))))...)). ( -34.60) >consensus ACCCA____________CUUCGUCCUUGGGAGCAAGGAUAGGAUCCUAGAUGGCUGCACCUGCUGACAGGUGUUGCCAGCAUUUUGCAUAUGUCUUCC_UCCAUUUCUGGCAACUGAGCA .....................(((((((....))))))).(((....(((((((.(((((((....))))))).))).((.....))....))))....))).................. (-29.23 = -30.23 + 1.00)

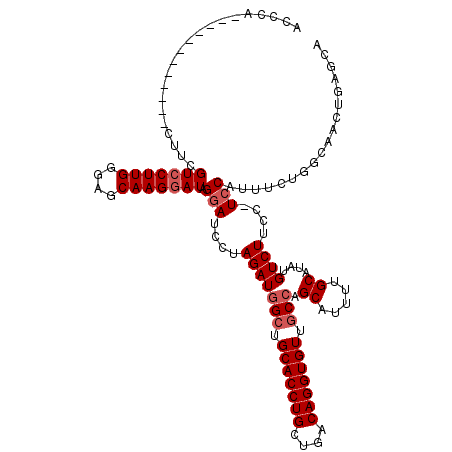
| Location | 3,305,495 – 3,305,602 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.02 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
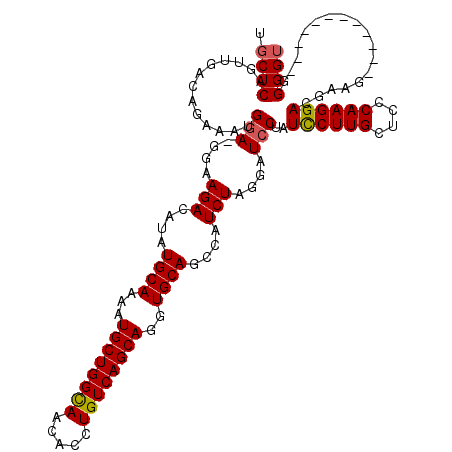
>3R_DroMel_CAF1 3305495 107 - 27905053 UGCUCAGUUGCCAGGAAUGGA-GGAAGACAUAUGCAAAAUGCUGGUAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGAUG------------GGGGU .((((.((((((((.((((..-......)))........).))))))))(((((....)))))....((((((((....)))((((((....)))))).))))------------))))) ( -35.00) >DroSec_CAF1 9553 107 - 1 UGCUCAGUUGACAGAAAUGGA-GGAAGACAUAUGCAAAAUGCUGGCAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUACUAUUCUUGGUCCCAAGGACGAAG------------UGGGU .(((((.((..((..(((((.-..........((((...((((((((.....))))))))..)))).((.....))...)))))..))((((...)))).)).------------))))) ( -32.10) >DroSim_CAF1 13162 107 - 1 UGCUCAGUUGACAGAAAUGGA-GGAAGACAUAUGCAAAAUGCUGGCAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGAAG------------UGGGU .(((((.(((.((....)).(-((((((....((((...((((((((.....))))))))..))))....)))....)))).((((((....)))))))))..------------))))) ( -35.00) >DroYak_CAF1 19775 120 - 1 UGCUCAGUUGCCAAAAAUGGAAGGAAGACAUAUGCAAAAUGCUGGCAACACCUGUCAGCAGGUGCAGCAAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGCAGGUACGUUUGGUGGGGGA .((......)).......((((((((((....((((...((((((((.....))))))))..))))....)))....)))).)))...((((((.(((((......)))))..)))))). ( -40.50) >consensus UGCUCAGUUGACAGAAAUGGA_GGAAGACAUAUGCAAAAUGCUGGCAACACCUGUCAGCAGGUGCAGCCAUCUAGGAUCCUAUCCUUGCUCCCAAGGACGAAG____________GGGGU .((((.............(((....(((....((((...((((((((.....))))))))..))))....)))....)))..((((((....))))))..................)))) (-27.90 = -28.02 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:19 2006