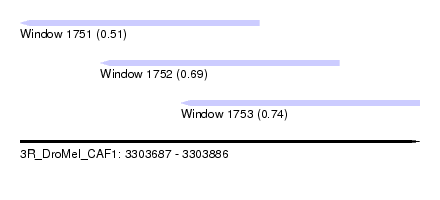
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,303,687 – 3,303,886 |
| Length | 199 |
| Max. P | 0.744059 |

| Location | 3,303,687 – 3,303,806 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3303687 119 - 27905053 CCU-CCCUUUAUUCGGAAGAAUAAAUCUCGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAUCGCCUUUGUAUGGCAUUGAAAAGUUCCGAUUGGCAAACUUAACAUGGCAAUGUUUAC ..(-((.(((((((....)))))))....)))((((((((((((.....))))))))(((.((((((.......)))(((....)))...))).)))..........))))......... ( -30.20) >DroSec_CAF1 7737 120 - 1 CCUUGCCUUUAUUCGGAAGAAUAAAUCUGGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCACGGCCUUUGUAUGGCAUUGAAAAGUUCCGAUUUGCAAACUUAACACGGCAAUGUUUAC ..((((((((((((....))))))).(((((.((..((((((((.....)))))))))))).)))..(((((((((.(((....))).)))...))))))........)))))....... ( -27.90) >DroSim_CAF1 6159 120 - 1 CCUUGCCUUUAUUCGGAAGAAUAAAUCUGGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAGCGCCUUUGUAUGGCAUUGAAAAGUUCCGAUUGGCAAACUUAACACGGCAAUGUUUAC ..((((((((((((....))))))).(((((.....((((((((.....)))))))).))))).(((...((.(((.(((....))).))))).)))...........)))))....... ( -32.60) >DroYak_CAF1 17951 118 - 1 CCUU--CUUUAUUCGGGAGAAUAAAUCACUGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAGCGCCUUUUUAUGGCAUUGAAAAGUUCCGAUUGACAAACUUAACACGGCGGUGUUUAC ....--.(((((((....))))))).......(((.((((((((.....))))))))((((((.(((.......))).)))..(((((..........))))).....))))))...... ( -22.60) >consensus CCUUGCCUUUAUUCGGAAGAAUAAAUCUCGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAGCGCCUUUGUAUGGCAUUGAAAAGUUCCGAUUGGCAAACUUAACACGGCAAUGUUUAC ..((((((((((((....)))))))....(((((..((((((((.....))))))))...((..(((.......)))..))....)))))..................)))))....... (-21.61 = -22.55 + 0.94)

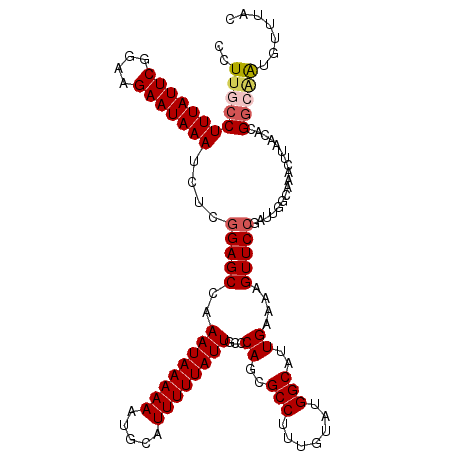
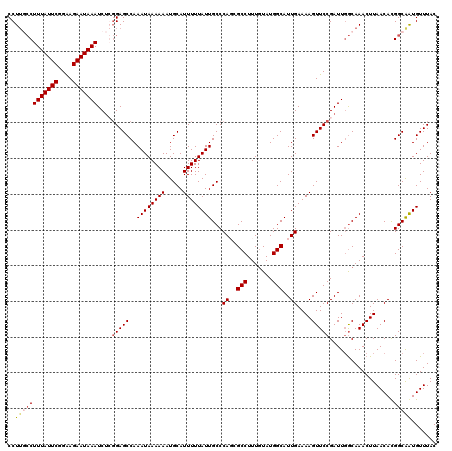
| Location | 3,303,727 – 3,303,846 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -24.92 |
| Energy contribution | -26.30 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3303727 119 - 27905053 AUCACGUAGUGUAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCU-CCCUUUAUUCGGAAGAAUAAAUCUCGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAUCGCCUUUGUAUGGCAUU .....((.((((((((.(((((((((.((..((((((....((-((.(((((((....)))))))....))))....))))))..)).)))))))))((.....)))))))))).))... ( -26.80) >DroSec_CAF1 7777 120 - 1 AUCACGUAGUGCAAAGAAAUAGAAACUCGCGUUUUGUCCCCCUUGCCUUUAUUCGGAAGAAUAAAUCUGGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCACGGCCUUUGUAUGGCAUU .....((.((((((((.....(((((....))))).........((((((((((....)))))))..((((.....((((((((.....)))))))).)))).))))))))))).))... ( -31.70) >DroSim_CAF1 6199 120 - 1 AUCACGUAGUGCAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCUUGCCUUUAUUCGGAAGAAUAAAUCUGGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAGCGCCUUUGUAUGGCAUU .....((.((((((((.(((((((((..(((((((..(((((.....(((((((....)))))))...))).)).......))))))))))))))))((.....)))))))))).))... ( -33.00) >DroYak_CAF1 17991 118 - 1 AUCACGUAGUGCAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCUU--CUUUAUUCGGGAGAAUAAAUCACUGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAGCGCCUUUUUAUGGCAUU .....(..(..(((((....(....).....)))))..)..)..--.(((((((....)))))))...(((.((..((((((((.....)))))))))).))).(((.......)))... ( -23.00) >consensus AUCACGUAGUGCAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCUUGCCUUUAUUCGGAAGAAUAAAUCUCGGAGCCAAAUAAAAAAUGCAUUUUUAUUGCCCAGCGCCUUUGUAUGGCAUU .....((.((((((((.(((((((((..(((((((..(((((.....(((((((....)))))))...))).)).......))))))))))))))))((.....)))))))))).))... (-24.92 = -26.30 + 1.38)

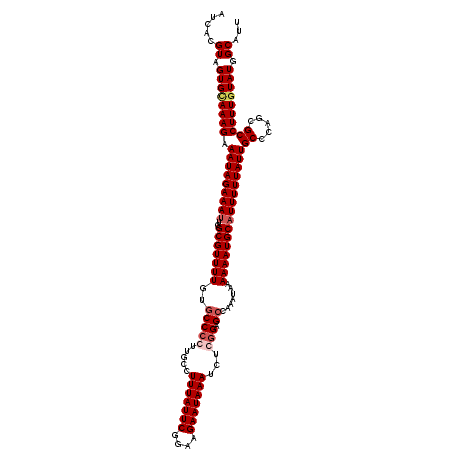
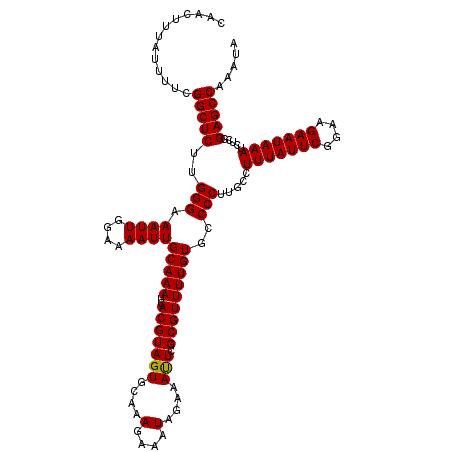
| Location | 3,303,767 – 3,303,886 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -26.84 |
| Energy contribution | -26.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3303767 119 - 27905053 CAACUUUAUUUUCGGCUCUUGGGAAAUUGGAAAAUUGCAAAUCACGUAGUGUAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCU-CCCUUUAUUCGGAAGAAUAAAUCUCGGAGCCAAAUA .............((((((.((((((((....))))(((....(((((((....(....)....))).))))....)))...)-)))(((((((....)))))))....))))))..... ( -29.50) >DroSec_CAF1 7817 120 - 1 CAACUUUAUUUUCGGCUCUUGGGAAAUUGGAAAAUUGCAAAUCACGUAGUGCAAAGAAAUAGAAACUCGCGUUUUGUCCCCCUUGCCUUUAUUCGGAAGAAUAAAUCUGGGAGCCAAAUA .............((((((..((.....((......(((((..(((((((....(....)....))).)))))))))........))(((((((....)))))))))..))))))..... ( -30.74) >DroSim_CAF1 6239 120 - 1 CAACUUUAUUUUCGGCUCUUGGGAAAUUGGAAAAUUGCAAAUCACGUAGUGCAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCUUGCCUUUAUUCGGAAGAAUAAAUCUGGGAGCCAAAUA .............((((((..((.((((....))))((((.....(..(..(((((....(....).....)))))..)..))))).(((((((....)))))))))..))))))..... ( -31.00) >DroYak_CAF1 18031 118 - 1 CAACUUUAUUUUCGGCUCUUGGGAAAUUGGAAAAUUGCAAAUCACGUAGUGCAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCUU--CUUUAUUCGGGAGAAUAAAUCACUGAGCCAAAUA .............(((((..(((.((((....))))............(..(((((....(....).....)))))..))))..--.(((((((....))))))).....)))))..... ( -26.30) >consensus CAACUUUAUUUUCGGCUCUUGGGAAAUUGGAAAAUUGCAAAUCACGUAGUGCAAAGAAAUAGAAAUUCGCGUUUUGUGCCCCUUGCCUUUAUUCGGAAGAAUAAAUCUCGGAGCCAAAUA .............(((((..(((.((((....))))(((((..(((((((....(....)....))).)))))))))..))).....(((((((....))))))).....)))))..... (-26.84 = -26.65 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:09 2006