| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 649,789 – 649,879 |
| Length | 90 |
| Max. P | 0.983319 |

| Location | 649,789 – 649,879 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
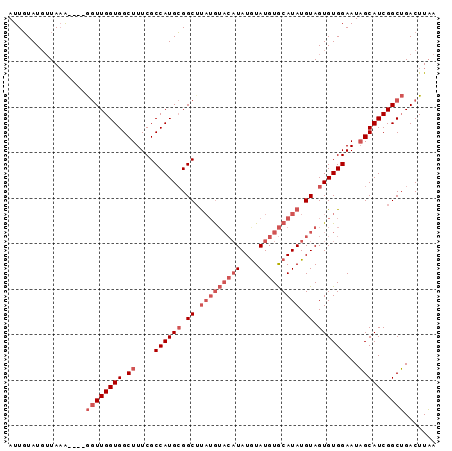
>3R_DroMel_CAF1 649789 90 + 27905053 UGAAGUCCGCCGAUGCGAUUCCACACUACAUAUGUACAUACAUA----CAUAAGCCUCAUGGCGAAAGCCACCAAGUCAGACGUAUCAUAAAAU ...........((((((.............((((((......))----)))).......((((....))))..........))))))....... ( -20.40) >DroSec_CAF1 70404 88 + 1 UUAAGUCAGCCGAUGCUAUUCCACACUACAUAUGCACAUACAUAUGUAU--GUGCCGCAUGGCGAAAGCCACCAACC----UUUAAGAUACAAU ....(((.....((((.....((((.((((((((......)))))))))--)))..))))(((....))).......----.....)))..... ( -23.70) >DroSim_CAF1 67371 90 + 1 UUAAGUCAGCCGAUGCUAUUCCACACUACAUAUGCACAUAUAUAUGUACAUAAGCCGCAUGGCGAAAGCCACCAACC----UUUAACAUACAAU ............((((..........((((((((......))))))))........))))(((....))).......----............. ( -15.17) >consensus UUAAGUCAGCCGAUGCUAUUCCACACUACAUAUGCACAUACAUAUGUACAUAAGCCGCAUGGCGAAAGCCACCAACC____UUUAACAUACAAU .......(((....))).........((((((((......))))))))...........((((....))))....................... (-13.80 = -14.47 + 0.67)

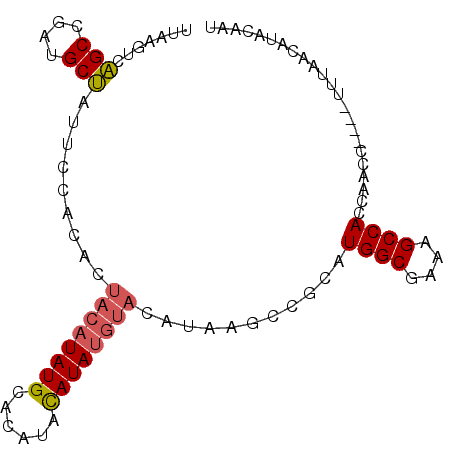
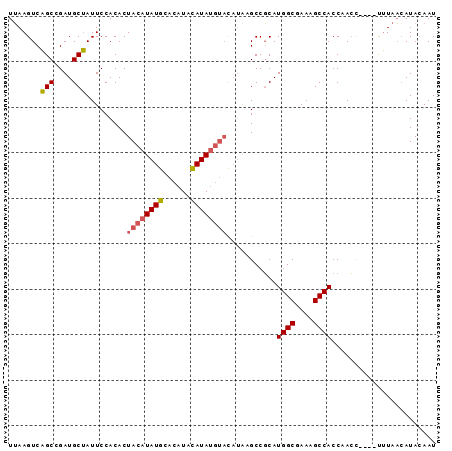
| Location | 649,789 – 649,879 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -16.53 |
| Energy contribution | -20.53 |
| Covariance contribution | 4.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 649789 90 - 27905053 AUUUUAUGAUACGUCUGACUUGGUGGCUUUCGCCAUGAGGCUUAUG----UAUGUAUGUACAUAUGUAGUGUGGAAUCGCAUCGGCGGACUUCA ......(((...(((((.(((.(((((....))))).)))..((((----(((((....)))))))))(((((....)))))...))))).))) ( -24.80) >DroSec_CAF1 70404 88 - 1 AUUGUAUCUUAAA----GGUUGGUGGCUUUCGCCAUGCGGCAC--AUACAUAUGUAUGUGCAUAUGUAGUGUGGAAUAGCAUCGGCUGACUUAA ............(----(((..((.(((....((((((.((((--((((....)))))))).......))))))...)))....))..)))).. ( -28.90) >DroSim_CAF1 67371 90 - 1 AUUGUAUGUUAAA----GGUUGGUGGCUUUCGCCAUGCGGCUUAUGUACAUAUAUAUGUGCAUAUGUAGUGUGGAAUAGCAUCGGCUGACUUAA ............(----(((..((.(((....((((((.((.(((((((((....))))))))).)).))))))...)))....))..)))).. ( -30.00) >consensus AUUGUAUGUUAAA____GGUUGGUGGCUUUCGCCAUGCGGCUUAUGUACAUAUGUAUGUGCAUAUGUAGUGUGGAAUAGCAUCGGCUGACUUAA .................((((((((.((....((((((.((.(((((((((....))))))))).)).))))))...))))))))))....... (-16.53 = -20.53 + 4.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:26 2006