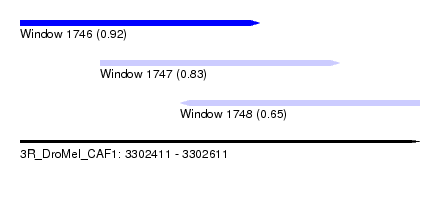
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,302,411 – 3,302,611 |
| Length | 200 |
| Max. P | 0.916202 |

| Location | 3,302,411 – 3,302,531 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
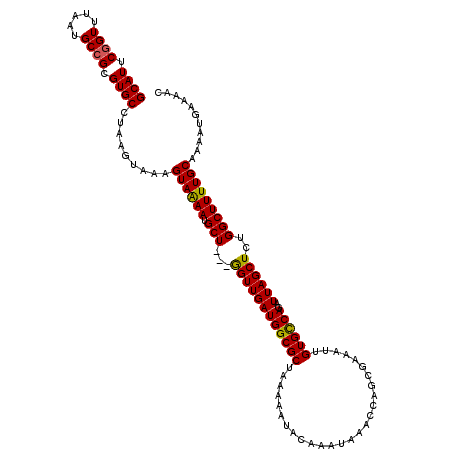
>3R_DroMel_CAF1 3302411 120 + 27905053 AAAGUGAACCAAAUAAUUCGCCACUAAUUUGAUUCUAAAUGUUAAAUGCACUAAUACGAAUUAGGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUU ...(((((........)))))..................((((....((.(((((....)))))))..))))..(((((((......(((.((....)).)))......))))))).... ( -23.00) >DroSec_CAF1 6583 120 + 1 AAAGUGAAUCAAAUAAUUCGCCACUAAUUUGAUUCUAAAUGUUAAAUGCACUAAUACGAAUUAAGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUU .....(((((((((............)))))))))....((((....((..((((....)))).))..))))..(((((((......(((.((....)).)))......))))))).... ( -22.50) >DroSim_CAF1 4931 120 + 1 AAAGUGAAUCAAAUAAUUCGCCACUAAUUUGAUUCUAAAUGUUAAAUGCACUAAUACGAAUUAAGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUU .....(((((((((............)))))))))....((((....((..((((....)))).))..))))..(((((((......(((.((....)).)))......))))))).... ( -22.50) >DroEre_CAF1 3617 120 + 1 AAAGUGAACUAGAUAAUUCGCCACUAAUUUGAUUCUAAAUGUUAAAUGCACUAAAACGAAUUAAGCGGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGGCACAAUU ((((((((.....(((((((......(((((((.......))))))).........)))))))(((..(((.....))).)))))))))))......((((((.....))))))...... ( -26.16) >DroYak_CAF1 9327 120 + 1 ACAGCGAACUAAAUAAUUCGCCACUAAUUUGAUUCUAAAUGUUAAAUGCACUAAUACGAAUUAAGCGGGACAAACUGUCAGGUUUCAUUUGGCAAAAGCCAGAGCUAAUCUGGCACAAUU ...(((((........)))))..((..((((((((....(((((.......))))).))))))))..))......(((((((((...((((((....))))))...)))))))))..... ( -29.40) >consensus AAAGUGAACCAAAUAAUUCGCCACUAAUUUGAUUCUAAAUGUUAAAUGCACUAAUACGAAUUAAGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUU ...(((((........)))))..((..((((((((....(((.....))).......))))))))..)).....(((((((.((...(((.((....)).)))...)).))))))).... (-20.46 = -20.10 + -0.36)

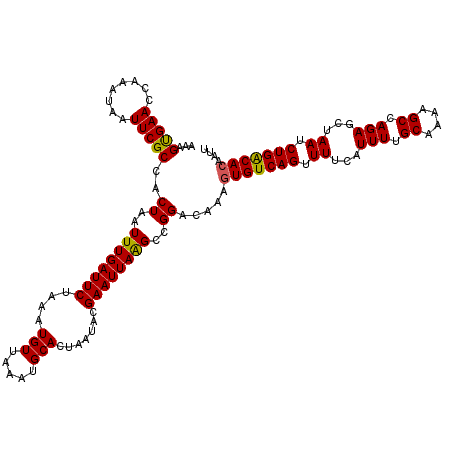
| Location | 3,302,451 – 3,302,571 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -21.26 |
| Energy contribution | -23.26 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3302451 120 + 27905053 GUUAAAUGCACUAAUACGAAUUAGGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUUUUGCUGGUUUAUUUGUAUUUUUAGCGCCAUCAACUAGCAG (((....((.(((((((((((.(((((((.(((((((((((......(((.((....)).)))......)))))))...)))))))))))))))))))...))).))....)))...... ( -32.90) >DroSec_CAF1 6623 95 + 1 GUUAAAUGCACUAAUACGAAUUAAGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUUUCGCUGGUUUAUUUG------------------------- ................(((((.(((((((.....(((((((......(((.((....)).)))......))))))).......))))))))))))------------------------- ( -22.50) >DroSim_CAF1 4971 120 + 1 GUUAAAUGCACUAAUACGAAUUAAGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUUUCGCUGGUUUAUUUGUAUAUUUAGCGCCAUCAACCAGCAG (((....((.(((((((((((.(((((((.....(((((((......(((.((....)).)))......))))))).......)))))))))))))))...))).))....)))...... ( -30.50) >DroEre_CAF1 3657 117 + 1 GUUAAAUGCACUAAAACGAAUUAAGCGGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGGCACAAUUUCGCAGGUUUAUUUGUAUUUUGAGCGCCAUCAACC---AG ((((((((((....((..((((..(((..((((((((........))))))).....((((((.....))))))....)..))).))))..)))))))))..)))..........---.. ( -26.20) >DroYak_CAF1 9367 117 + 1 GUUAAAUGCACUAAUACGAAUUAAGCGGGACAAACUGUCAGGUUUCAUUUGGCAAAAGCCAGAGCUAAUCUGGCACAAUUUCGCUGGUUUAUUUGUAUUUUUAGCGCCAUCAACC---AG (((....((.(((((((((((..((((..(.....(((((((((...((((((....))))))...)))))))))...)..)))).....))))))))...))).))....))).---.. ( -35.70) >consensus GUUAAAUGCACUAAUACGAAUUAAGCCGGACAAAGUGUCAGUUUUCAUUUUGCAAAAGCCAGAGCUAAUCUGACACAAUUUCGCUGGUUUAUUUGUAUUUUUAGCGCCAUCAACC___AG .......((...(((((((((.(((((((.....(((((((.((...(((.((....)).)))...)).))))))).......))))))))))))))))....))............... (-21.26 = -23.26 + 2.00)


| Location | 3,302,491 – 3,302,611 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -27.13 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3302491 120 - 27905053 GCAUUCGGUUUAAUGCCGCGUGCCUAAGUAAAGUAAAAUGCUGCUAGUUGAUGGCGCUAAAAAUACAAAUAAACCAGCAAAAUUGUGUCAGAUUAGCUCUGGCUUUUGCAAAAUGAAAAC ((((.((((.....)))).)))).........((((((.((((((((((..((((((...........................)))))))))))))...)))))))))........... ( -28.93) >DroSim_CAF1 5011 120 - 1 GCAUUCGGUUUAAUGCCGCGUGCCUAAGUAAAGUAAAAUGCUGCUGGUUGAUGGCGCUAAAUAUACAAAUAAACCAGCGAAAUUGUGUCAGAUUAGCUCUGGCUUUUGCAAAAUGAAAAC ((((.((((.....)))).))))....((((((...(((..((((((((..((............))....))))))))..)))..((((((.....))))))))))))........... ( -30.70) >DroEre_CAF1 3697 117 - 1 GCAUUCGGUUUAAUGCGGCGUGCCUAAGUAAAGUAAAAUGCU---GGUUGAUGGCGCUCAAAAUACAAAUAAACCUGCGAAAUUGUGCCAGAUUAGCUCUGGCUUUUGCAAAAUGAAAAC ...(((((((((((...(.(((((.......(((.....)))---.......))))).)...)).....))))))(((((((....((((((.....)))))))))))))....)))... ( -29.34) >DroYak_CAF1 9407 117 - 1 GCAUUCGGUUUAAUGCCGCGUGCCUAAGUAAAGUAGAAUGCU---GGUUGAUGGCGCUAAAAAUACAAAUAAACCAGCGAAAUUGUGCCAGAUUAGCUCUGGCUUUUGCCAAAUGAAACC ((((.((((.....)))).)))).........((((((((((---((((..((............))....)))))))).......((((((.....))))))))))))........... ( -32.60) >consensus GCAUUCGGUUUAAUGCCGCGUGCCUAAGUAAAGUAAAAUGCU___GGUUGAUGGCGCUAAAAAUACAAAUAAACCAGCGAAAUUGUGCCAGAUUAGCUCUGGCUUUUGCAAAAUGAAAAC ((((.((((.....)))).)))).........((((((.(((...((((((((((((...........................))))))..))))))..)))))))))........... (-27.13 = -26.75 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:04 2006