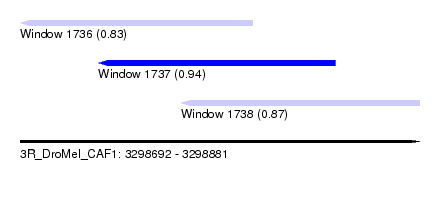
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,298,692 – 3,298,881 |
| Length | 189 |
| Max. P | 0.939144 |

| Location | 3,298,692 – 3,298,802 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -18.65 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3298692 110 - 27905053 ACCAACAGCCAGGGAU--CAAGACUAUUAUCUGUCAUUGAGUCCUAGACAGAUAGUUUAGUCUCUGGUUUACAGAACACCCAAAUGGAAAUGAAACAC-GCGCUUUUGUCGAC ..(((.((((.(((..--...((((((((((((((...........))))))))))..))))((((.....))))...)))..((....)).......-).))).)))..... ( -27.60) >DroVir_CAF1 1057 107 - 1 AGCAA-UGCCAUUUAAUGCUAGACUGUUAUUUGUCAUUAAG-UCAAGACAAAUGAUUUAGGCUUGGGCGUACAAUC-UUUAU-UUUUUAUUGAAAC--AGUGCUAUUGUCGCC ..(((-.(((.((((....))))..((((((((((......-....))))))))))...))))))((((.(((((.-..(((-(.(((....))).--))))..))))))))) ( -21.70) >DroSec_CAF1 3143 109 - 1 ACCAAUAGCCAGGGAU--CAAGACUAUUAUCUGUCCUUAAGUUCUAGACAGGUAGUUUAGUCUCUGUCUUACAGAACACCCAAAU-GAAAUGAAACAC-GCGCUAUUGUCGAC ..((((((((.(((..--...((((((((((((((...........))))))))))..))))((((.....))))...)))...(-(........)).-).)))))))..... ( -29.10) >DroSim_CAF1 1543 109 - 1 ACCAACAGCCAGGGAU--CAAGACUAUUAUCUGUCCUUAAGUCCUAGACAGAUAGUUUAGUCUCUGGCUUACAGAACACCCAAAA-GAAAUGAAACAC-GCGCUAUUGUCGAC ..(((.((((.(((..--...((((((((((((((...........))))))))))..))))((((.....))))...)))...(-....).......-).))).)))..... ( -26.20) >DroEre_CAF1 500 110 - 1 ACCACAUGCCAGGUAU--CAAGACUAUUAUCUGUCCUUAAGUCCUAGACAGAUAGUUUAGUUUCUGGCUUACAGAACACCCAACUAGAAAUGAAACAC-GCACUAUUGUCGAC (((........)))..--...(((...((((((((...........))))))))((((.((((((((................)))))))).))))..-........)))... ( -22.29) >DroYak_CAF1 6270 110 - 1 ACCAAAAGCCAGGUAU--CAAGACUAUUGUCUGUCCUUAAGUCCUAGACAGAUAGUUUAGUUUCUGGCUUACAGAACACCAAAAUAAAAAUGAAACAC-GCACUUUUGUUGAC .....((((((((...--(.(((((((((((((..(....)..).))))).))))))).)..))))))))............(((((((.((......-.)).)))))))... ( -25.40) >consensus ACCAAAAGCCAGGGAU__CAAGACUAUUAUCUGUCCUUAAGUCCUAGACAGAUAGUUUAGUCUCUGGCUUACAGAACACCCAAAU_GAAAUGAAACAC_GCGCUAUUGUCGAC ......((((((........(((((((((((((((...........))))))))))..)))))))))))............................................ (-18.65 = -17.93 + -0.72)

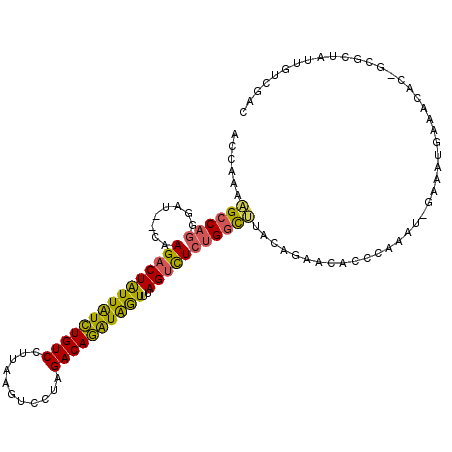

| Location | 3,298,729 – 3,298,841 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -22.54 |
| Energy contribution | -20.60 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3298729 112 - 27905053 GCACUCCAGAAU-UUGCAGUGGCCGUAAGGCCAGCGGGACACCAACAGCCAGGGAU--CAAGACUAUUAUCUGUCAUUGAGUCCUAGACAGAUAGUUUAGUCUCUGGUUUACAGA (((((.(((...-))).)))((((....)))).))((....))...((((((((((--..((((((...((((((...........)))))))))))).))))))))))...... ( -38.70) >DroVir_CAF1 1091 113 - 1 GGCUCAGAGCAUGUAGCGGUUGCUGUAAGGCCAACGGAACAGCAA-UGCCAUUUAAUGCUAGACUGUUAUUUGUCAUUAAG-UCAAGACAAAUGAUUUAGGCUUGGGCGUACAAU .(((((((((((.....(((((((((..(.....)...)))))).-.))).....)))))...((((((((((((......-....))))))))))..))..))))))....... ( -31.60) >DroSec_CAF1 3179 112 - 1 GCACUCCAGACU-UUGCAGUGGCCGUAAGGCCAGCGGGACACCAAUAGCCAGGGAU--CAAGACUAUUAUCUGUCCUUAAGUUCUAGACAGGUAGUUUAGUCUCUGUCUUACAGA ((..........-..((..(((((....)))))))((....))....))..(((((--..(((((((((((((((...........))))))))))..)))))..)))))..... ( -35.40) >DroEre_CAF1 537 112 - 1 GCACUCCAGAAU-UGGCAGUAGCCGUAAGGCCAGCGGGACACCACAUGCCAGGUAU--CAAGACUAUUAUCUGUCCUUAAGUCCUAGACAGAUAGUUUAGUUUCUGGCUUACAGA .....(((((.(-(((((((.(((....)))..))((....))...))))))....--...((((((((((((((...........))))))))))..)))))))))........ ( -35.00) >DroYak_CAF1 6307 112 - 1 GCCCUCCAGAAU-UGGCAGUGGCCGUAAGGCCAGCGGAACACCAAAAGCCAGGUAU--CAAGACUAUUGUCUGUCCUUAAGUCCUAGACAGAUAGUUUAGUUUCUGGCUUACAGA ((.((((.....-.)).))(((((....)))))))((....))..((((((((...--(.(((((((((((((..(....)..).))))).))))))).)..))))))))..... ( -37.90) >DroMoj_CAF1 722 113 - 1 GCCUCGGAGUAUGCGGCGGUUGCUGUAAGGCCAACGGAACAGCAA-UGCCAUUUAACGCUAGACUGUUGUUUGCCAUCGAG-UCAAGGCAGAUGAUUUAGGCGUGGGCGCACAAU ((((..........((((.(((((((..(.....)...)))))))-)))).....(((((.....((..((((((......-....))))))..))...)))))))))....... ( -37.30) >consensus GCACUCCAGAAU_UGGCAGUGGCCGUAAGGCCAGCGGAACACCAA_AGCCAGGUAU__CAAGACUAUUAUCUGUCAUUAAGUCCUAGACAGAUAGUUUAGUCUCUGGCUUACAGA ..................((((((....(((....((....))....)))...........((((((((((((((...........))))))))))..))))...)))).))... (-22.54 = -20.60 + -1.94)

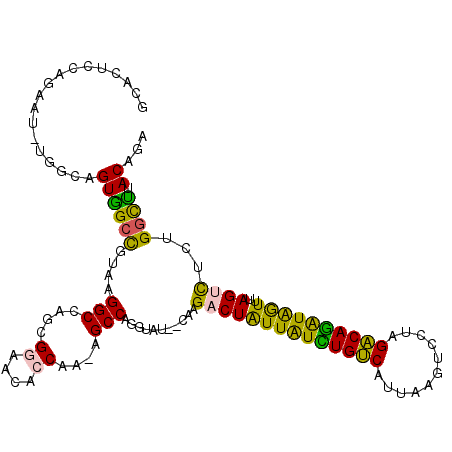
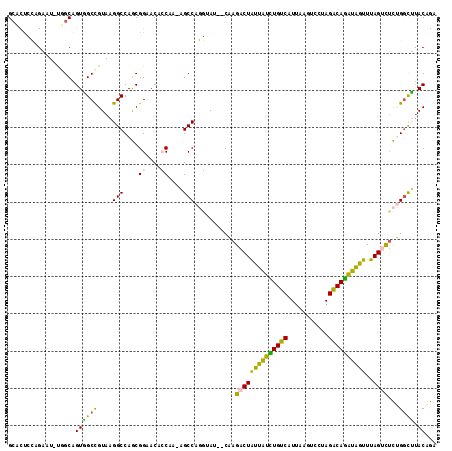
| Location | 3,298,768 – 3,298,881 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -25.18 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3298768 113 - 27905053 UCUAGCAAAUCUAGUUUCAUCCGUUGUAAUCUUUGCUGCUGCACUCCAGAAU-UUGCAGUGGCCGUAAGGCCAGCGGGACACCAAC---AGCCAGG-GAU--CAAGACUAUUAUCUGUCA ...........((((((.(((((((((.......(((((((((.........-.))))))((((....)))))))((....)).))---)))...)-)))--..)))))).......... ( -32.70) >DroVir_CAF1 1129 113 - 1 AAUAGCACA--CAGUUUCGUUCGUUGUAAUCUUAGCGGCUGGCUCAGAGCAUGUAGCGGUUGCUGUAAGGCCAACGGAACAGCAA----UGCCAUU-UAAUGCUAGACUGUUAUUUGUCA ..(((((..--..((((((((.(((.......(((((((((.((((.....)).)))))))))))...))).)))))))).((..----.))....-...)))))(((........))). ( -30.20) >DroGri_CAF1 1485 118 - 1 AAAAGCACA--CAGUUUCGUCCGUUGUAAUCUUAGCGGCCGUCUCGGAGCAUGCGGCGGUCGCUGUAAGGCCAGCGGAACAACAAACGCUGCCAUUUUUACGCUAGACUGUUGUUUGUCG ..(((((.(--((((((.((.....((((...(((((((((((.((.....)).)))))))))))...(((.((((..........)))))))....)))))).)))))))))))).... ( -41.50) >DroEre_CAF1 576 113 - 1 UUUAGCAAAUUUAGUUUCAUCCGUUGUAAUCUUUGCUGCUGCACUCCAGAAU-UGGCAGUAGCCGUAAGGCCAGCGGGACACCACA---UGCCAGG-UAU--CAAGACUAUUAUCUGUCC ....(((.((.((((((...((((((.......(((((((............-.)))))))(((....)))))))))((.(((...---.....))-).)--).))))))..)).))).. ( -30.32) >DroYak_CAF1 6346 113 - 1 UCCAGCAAAUUUUGUUUCAUCCGUUGUAAUCUUUGCUGCUGCCCUCCAGAAU-UGGCAGUGGCCGUAAGGCCAGCGGAACACCAAA---AGCCAGG-UAU--CAAGACUAUUGUCUGUCC ..((((((.(((((.....((((((............((((((.........-.))))))((((....))))))))))..(((...---.....))-)..--)))))...))).)))... ( -32.60) >DroMoj_CAF1 760 113 - 1 AAUAGCACA--AGGUUUCGUCCGCUGUAAUCUUAGCGGCCGCCUCGGAGUAUGCGGCGGUUGCUGUAAGGCCAACGGAACAGCAA----UGCCAUU-UAACGCUAGACUGUUGUUUGCCA ....(((..--..(((((((..(((.......(((((((((((.((.....)).)))))))))))...)))..))))))).....----)))....-....((.((((....)))))).. ( -37.40) >consensus AAUAGCAAA__UAGUUUCAUCCGUUGUAAUCUUAGCGGCUGCCCCCCAGAAU_UGGCAGUGGCCGUAAGGCCAGCGGAACACCAAA___UGCCAGG_UAA__CAAGACUAUUAUCUGUCA ....(((............(((((((...((((.(((((((((...........)))))))))...)))).)))))))...........))).............(((........))). (-25.18 = -24.52 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:54 2006