
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,289,128 – 3,289,239 |
| Length | 111 |
| Max. P | 0.528870 |

| Location | 3,289,128 – 3,289,239 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -27.80 |
| Energy contribution | -29.25 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3289128 111 + 27905053 AAUGUCAAACGGAAACAAGUUUCCAAGUCAGACUGCAUGAUGGAGCACACAUGUCCUGGCGGCCACAGGACUC---CUAGC---AAGCUAACAC-UCCUUCGA--CCAGUCGGGCGACAA ..((((....(((((....)))))..(((.(((((..(((.((((.......((((((.......))))))..---.(((.---...)))...)-))).))).--.))))).))))))). ( -39.10) >DroSec_CAF1 42089 111 + 1 AAUGUCAAACGGAAACAAGUUUCCAAGUCAGACUGCAUGAUGGAGCACACAUGUCCUGGCGGCCACAGGACUC---CUAGC---AGGCUAACAC-UCCUUCGA--CCAGUCGGACGACAA ..((((....(((((....)))))..(((.(((((..(((.((((.......((((((.......))))))..---.(((.---...)))...)-))).))).--.))))).))))))). ( -39.70) >DroSim_CAF1 43268 111 + 1 AAUGUCAAACGGAAACAAGUUUCCAAGUCAGACUGCAUGAUGGAGCACACAUGUCCUGGCGGCCACAGGACUC---CUAGC---AGGCUAACAC-UCCUUCGA--CCAGUCGGGCGACAA ..((((....(((((....)))))..(((.(((((..(((.((((.......((((((.......))))))..---.(((.---...)))...)-))).))).--.))))).))))))). ( -39.10) >DroEre_CAF1 43818 108 + 1 AAUGUCAAACGGAAACAAGUUUCCAAGUCAGACUGCAUGAUGGAGCACACAUGUCCUGGCGGCCACAGGACUC---CUA------GGCUUCCAC-UCCUUCGA--CCAGUCGGGCGACAA ..((((....(((((....)))))..(((.(((((..(((.((((.......((((((.......))))))..---...------((...)).)-))).))).--.))))).))))))). ( -38.00) >DroWil_CAF1 153118 115 + 1 AAUGUCAAACGGAAACAAGUUGCCAAGUCAGCUUGGUUGAUGGAGGA--CAUGUCCUG---GUCACAGGACACAACCAACCACUGAGCCUGCGCUUGCUUCAAGUGCAGUCGAGCAACAA ..........(....)..((((((...((((.((((((((((.....--)))((((((---....)))))).)))))))...))))(.(((((((((...))))))))).)).))))).. ( -45.00) >DroYak_CAF1 61508 111 + 1 AAUGUCAAACGGAAACAAGUUUCCAAGUCAGACUGCAUGAUGGAGCACACACGUCCUGGCGGUCACAGGACUC---CUAGC---AGGCUUCCAC-UCCUUCGA--CCACUCGGGCGACAA ..((((....(((((....)))))..(((.((.((..(((.((((.......((((((.......)))))).(---(....---.))......)-))).))).--.)).)).))))))). ( -32.70) >consensus AAUGUCAAACGGAAACAAGUUUCCAAGUCAGACUGCAUGAUGGAGCACACAUGUCCUGGCGGCCACAGGACUC___CUAGC___AGGCUAACAC_UCCUUCGA__CCAGUCGGGCGACAA ..((((....(((((....)))))..(((.(((((..(((.(((........((((((.......))))))......((((.....)))).....))).)))....))))).))))))). (-27.80 = -29.25 + 1.45)

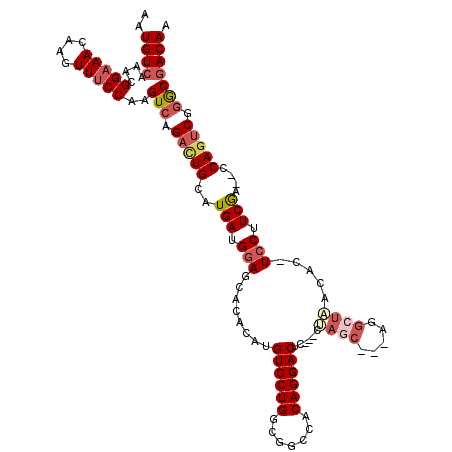
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:41 2006