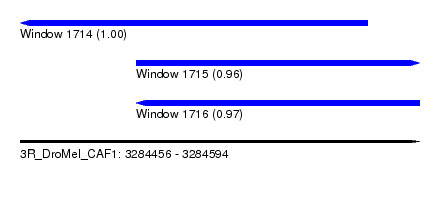
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,284,456 – 3,284,594 |
| Length | 138 |
| Max. P | 0.995236 |

| Location | 3,284,456 – 3,284,576 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.94 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
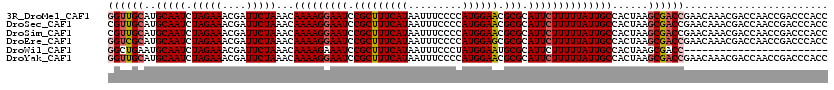
>3R_DroMel_CAF1 3284456 120 - 27905053 GGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAACAAACGACCAACCGACCCACC ((((((..(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))))))))......)))))).......((......))....... ( -27.70) >DroSec_CAF1 37533 120 - 1 CGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAACAAACGACCAACCGACCCACC .(((((..(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))))))))......)))))........((......))....... ( -23.80) >DroSim_CAF1 38739 120 - 1 CGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAACAAACGACCAACCGACCCACC .(((((..(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))))))))......)))))........((......))....... ( -23.80) >DroEre_CAF1 39115 120 - 1 GGUCGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAGCGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAACAAACGACCAACCGACCCACC ((((((..(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))))))))......)))))).......((......))....... ( -29.50) >DroWil_CAF1 147259 96 - 1 GGCUGAAUGCAAUCUAGAAACGAUUCUAAACAAAAGAAAUCCGCUUUCAUAAUUUCCCUAUGGAAUGCGCAUUCUUUUUAUUGCCACUAAGCGACC------------------------ .(((....(((((.(((((....)))))...(((((((...((((((((((.......))))))).)))..))))))).))))).....)))....------------------------ ( -21.10) >DroYak_CAF1 56867 120 - 1 GGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAACAAACGACCAACCGACCCACC ((((((..(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))))))))......)))))).......((......))....... ( -27.70) >consensus GGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAACAAACGACCAACCGACCCACC ((((((..(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))))))))......))))))........................ (-23.21 = -23.77 + 0.56)

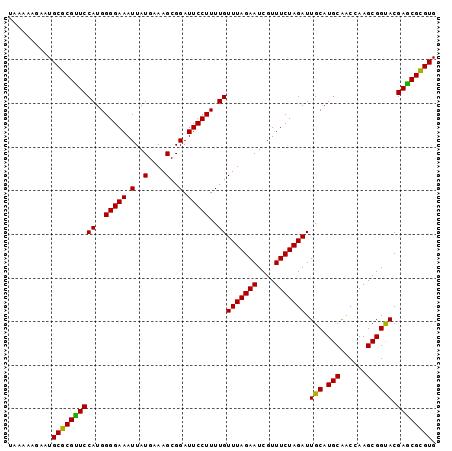
| Location | 3,284,496 – 3,284,594 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -26.26 |
| Energy contribution | -27.46 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3284496 98 + 27905053 UAAAAAGAAUGCGCGUUCCAUGGGGAAAUUAUGAAAGCGGAUUCCUUUUGUUUAGAAUCGUUUCUAGAUUGCAUGCAACCAAGCGGUACGAGCGCGUG ..........(((((((((((((((((.(..(....)..).)))))...((((((((....))))))))..))))..(((....)))..)))))))). ( -27.70) >DroSec_CAF1 37573 98 + 1 UAAAAAGAAUGCGCGUUCCAUGGGGAAAUUAUGAAAGCGGAUUCCUUUUGUUUAGAAUCGUUUCUAGAUUGCAUGCAACGAAGCGGCACGAGCGCGUG ..........((((((((((..(((((.(..(....)..).)))))..))(((((((....))))))).(((.(((......)))))).)))))))). ( -28.80) >DroSim_CAF1 38779 98 + 1 UAAAAAGAAUGCGCGUUCCAUGGGGAAAUUAUGAAAGCGGAUUCCUUUUGUUUAGAAUCGUUUCUAGAUUGCAUGCAACGAAGCGGCACGAGCGCGUG ..........((((((((((..(((((.(..(....)..).)))))..))(((((((....))))))).(((.(((......)))))).)))))))). ( -28.80) >DroEre_CAF1 39155 98 + 1 UAAAAAGAAUGCGCGCUCCAUGGGGAAAUUAUGAAAGCGGAUUCCUUUUGUUUAGAAUCGUUUCUAGAUUGCAUGCGACCAAGCGGUACGAGCGUGUG ..........(((((((((((((((((.(..(....)..).)))))...((((((((....))))))))..))))..(((....)))..)))))))). ( -28.30) >DroYak_CAF1 56907 98 + 1 UAAAAAGAAUGCGCGUUCCAUGGGGAAAUUAUGAAAGCGGAUUCCUUUUGUUUAGAAUCGUUUCUAGAUUGCAUGCAACCAAGCGGUACGAACGUGUG ..........(((((((((((((((((.(..(....)..).)))))...((((((((....))))))))..))))..(((....)))..)))))))). ( -25.80) >consensus UAAAAAGAAUGCGCGUUCCAUGGGGAAAUUAUGAAAGCGGAUUCCUUUUGUUUAGAAUCGUUUCUAGAUUGCAUGCAACCAAGCGGUACGAGCGCGUG ..........((((((((((..(((((.(..(....)..).)))))..))(((((((....))))))).(((.(((......)))))).)))))))). (-26.26 = -27.46 + 1.20)

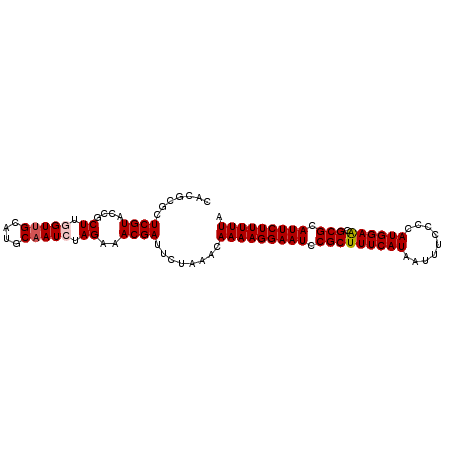
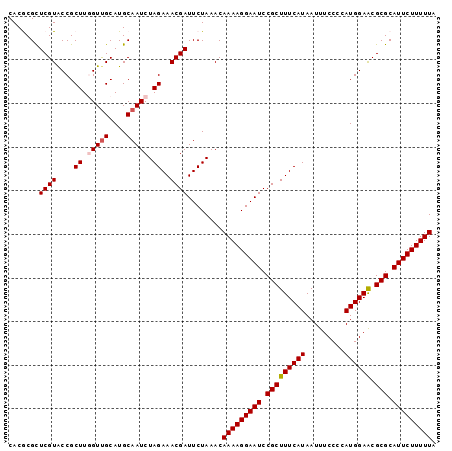
| Location | 3,284,496 – 3,284,594 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
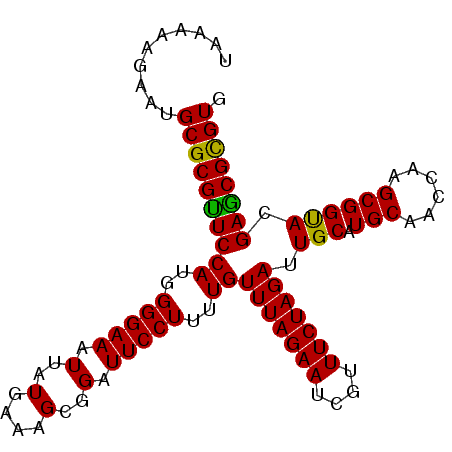
>3R_DroMel_CAF1 3284496 98 - 27905053 CACGCGCUCGUACCGCUUGGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUA .......((((....((.(((((....))))).))..))))........(((((((((.(((((((((.........)))))).))).))))))))). ( -22.40) >DroSec_CAF1 37573 98 - 1 CACGCGCUCGUGCCGCUUCGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUA ...((((....)).((......))..))....(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))). ( -21.30) >DroSim_CAF1 38779 98 - 1 CACGCGCUCGUGCCGCUUCGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUA ...((((....)).((......))..))....(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))). ( -21.30) >DroEre_CAF1 39155 98 - 1 CACACGCUCGUACCGCUUGGUCGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAGCGCGCAUUCUUUUUA .....((.((.(((....)))))...))....(((((....)))))...(((((((((.(((((((((.........)))))).))).))))))))). ( -21.00) >DroYak_CAF1 56907 98 - 1 CACACGUUCGUACCGCUUGGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUA .......((((....((.(((((....))))).))..))))........(((((((((.(((((((((.........)))))).))).))))))))). ( -22.50) >consensus CACGCGCUCGUACCGCUUGGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUA .......((((....((.(((((....))))).))..))))........(((((((((.(((((((((.........)))))).))).))))))))). (-20.92 = -21.36 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:33 2006