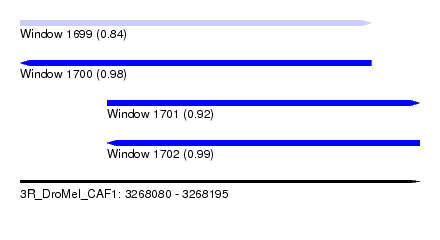
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,268,080 – 3,268,195 |
| Length | 115 |
| Max. P | 0.994364 |

| Location | 3,268,080 – 3,268,181 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -15.92 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3268080 101 + 27905053 ACCCGGCACACAAAUCCAAGCCCAAAACCAGGCUCAAACCGAACUCAGCUCCAAAUUUCAAGCGUAAACCCAAACCCAUCGCUUGACAUUUGCUCAACAUG ....(((...........((((........))))......(....).))).(((((.(((((((...............))))))).)))))......... ( -17.16) >DroSec_CAF1 21278 101 + 1 ACCCGGCACACAAAUCCAAGCCCAAAACCUGGCUCAAGCCGAACUCGGUUCCGAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUG ...((((...........((((........))))...)))).....((((.(((((.(((((((((..........)).))))))).)))))...)))).. ( -26.34) >DroSim_CAF1 22160 101 + 1 ACCCGGCACACAAAUCCAAGCCCAAAACCAGGCUCAAACCGAACUCGGUUCCGAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUG ....(((............)))......((((....(((((....))))).(((((.(((((((((..........)).))))))).))))).....)))) ( -26.50) >DroYak_CAF1 40724 100 + 1 ACCCA-UACACAAGUCCAAACCCAAAACCAGGCUUAAACCGAACUCGGCUUCAAAUUUCAAGCGUGAACUCGAACCCAUCGCUUGACAUUCGCUCAACCUG .....-......................((((......(((....))).........(((((((((..........)).)))))))...........)))) ( -16.20) >consensus ACCCGGCACACAAAUCCAAGCCCAAAACCAGGCUCAAACCGAACUCGGCUCCAAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUG ..................((((........))))..(((((....))))).(((((.(((((((...............))))))).)))))......... (-15.92 = -16.86 + 0.94)

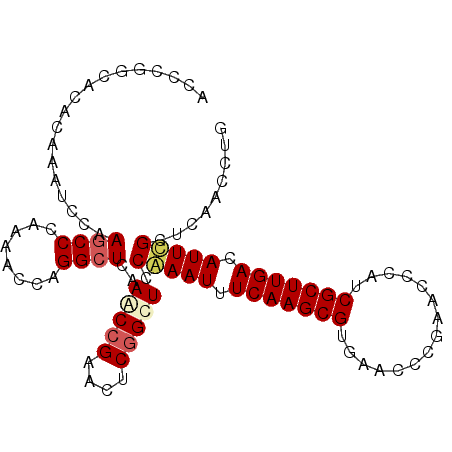
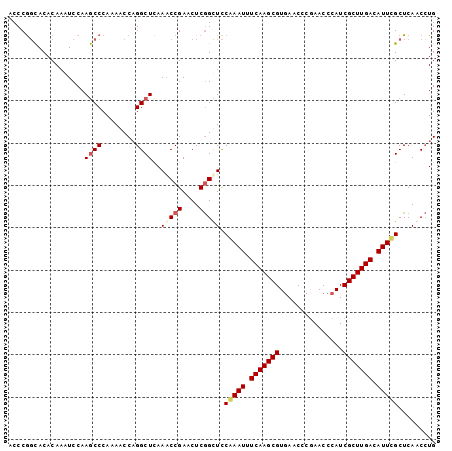
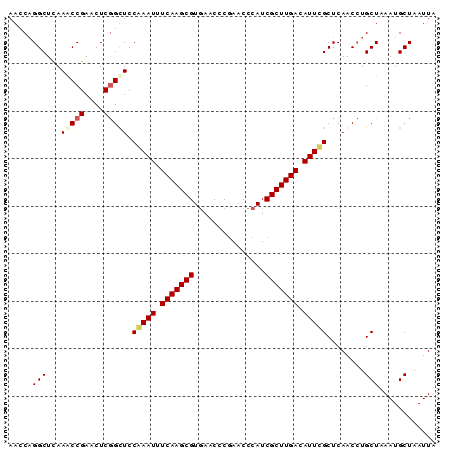
| Location | 3,268,080 – 3,268,181 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -32.34 |
| Energy contribution | -31.40 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3268080 101 - 27905053 CAUGUUGAGCAAAUGUCAAGCGAUGGGUUUGGGUUUACGCUUGAAAUUUGGAGCUGAGUUCGGUUUGAGCCUGGUUUUGGGCUUGGAUUUGUGUGCCGGGU .........(((((.(((((((.((..(....)..))))))))).)))))((((((....))))))..(((((((...(.((........)).)))))))) ( -28.60) >DroSec_CAF1 21278 101 - 1 CAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGCUUGAGCCAGGUUUUGGGCUUGGAUUUGUGUGCCGGGU ..((((...(((((.(((((((.((..(....)..))))))))).))))).))))..(..((((.((((((((((.....)))))).))))...))))..) ( -37.80) >DroSim_CAF1 22160 101 - 1 CAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGUUUGAGCCUGGUUUUGGGCUUGGAUUUGUGUGCCGGGU ..(((...((((((.((((((..(..(..((((((((.(((.(((.((((....)))))))))).))))))))..)..).))))))))))))..))).... ( -38.90) >DroYak_CAF1 40724 100 - 1 CAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGAGUUCACGCUUGAAAUUUGAAGCCGAGUUCGGUUUAAGCCUGGUUUUGGGUUUGGACUUGUGUA-UGGGU ..((((...(((((.(((((((.((..(....)..))))))))).))))).))))......(((((((((((......)))))))))))......-..... ( -33.70) >consensus CAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGUUUGAGCCUGGUUUUGGGCUUGGAUUUGUGUGCCGGGU ((((((...(((((.(((((((.((..(....)..))))))))).)))))((((((....))))))((((((......)))))).)))))).......... (-32.34 = -31.40 + -0.94)

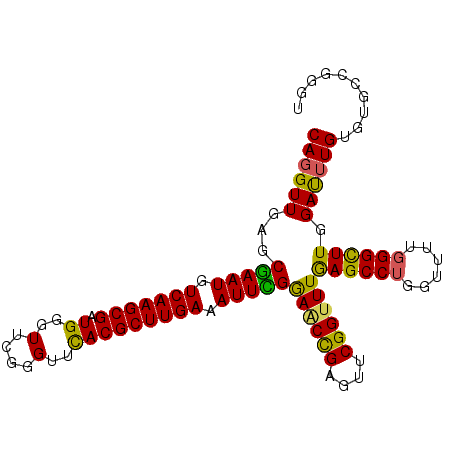
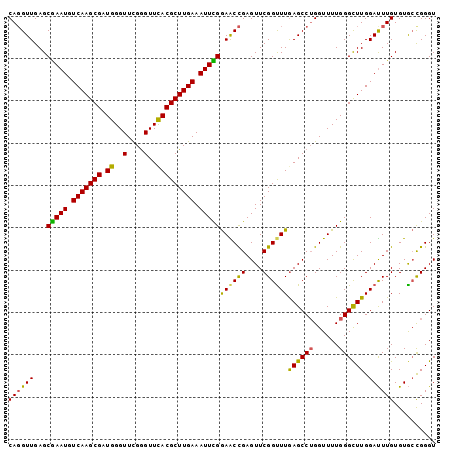
| Location | 3,268,105 – 3,268,195 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.76 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3268105 90 + 27905053 AACCAGGCUCAAACCGAACUCAGCUCCAAAUUUCAAGCGUAAACCCAAACCCAUCGCUUGACAUUUGCUCAACAUGCUAAAUGCUAAUUA .....((......))...........(((((.(((((((...............))))))).)))))........((.....))...... ( -13.76) >DroSec_CAF1 21303 90 + 1 AACCUGGCUCAAGCCGAACUCGGUUCCGAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUA ....((((....)))).....((((.(((((.(((((((((..........)).))))))).)))))...)))).((.....))...... ( -24.00) >DroSim_CAF1 22185 90 + 1 AACCAGGCUCAAACCGAACUCGGUUCCGAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUA ...((((....(((((....))))).(((((.(((((((((..........)).))))))).))))).....)))).............. ( -24.30) >DroYak_CAF1 40748 90 + 1 AACCAGGCUUAAACCGAACUCGGCUUCAAAUUUCAAGCGUGAACUCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUA ...((((......(((....))).........(((((((((..........)).)))))))...........)))).............. ( -16.60) >consensus AACCAGGCUCAAACCGAACUCGGCUCCAAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUA .....(((...(((((....))))).(((((.(((((((...............))))))).)))))........)))............ (-16.07 = -16.76 + 0.69)

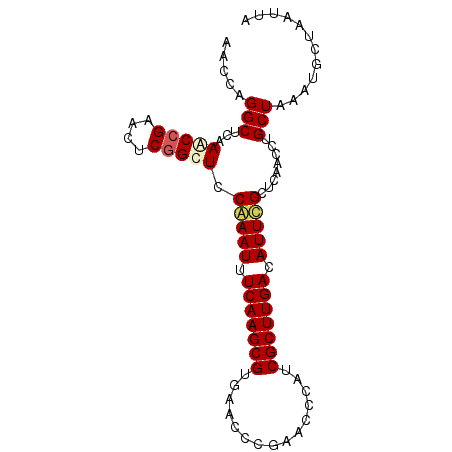
| Location | 3,268,105 – 3,268,195 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.15 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3268105 90 - 27905053 UAAUUAGCAUUUAGCAUGUUGAGCAAAUGUCAAGCGAUGGGUUUGGGUUUACGCUUGAAAUUUGGAGCUGAGUUCGGUUUGAGCCUGGUU ...(((((((.....))))))).(((((.(((((((.((..(....)..))))))))).))))).((((..((((.....))))..)))) ( -23.00) >DroSec_CAF1 21303 90 - 1 UAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGCUUGAGCCAGGUU .....(((......((((((((((((((.(((((((.((..(....)..))))))))).))))(....)..))))))))))......))) ( -30.20) >DroSim_CAF1 22185 90 - 1 UAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGUUUGAGCCUGGUU ..............((((((...(((((.(((((((.((..(....)..))))))))).)))))((((((....)))))).))))))... ( -33.80) >DroYak_CAF1 40748 90 - 1 UAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGAGUUCACGCUUGAAAUUUGAAGCCGAGUUCGGUUUAAGCCUGGUU ..............((((((...(((((.(((((((.((..(....)..))))))))).)))))((((((....)))))).))))))... ( -31.00) >consensus UAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGUUUGAGCCUGGUU ..............((((((...(((((.(((((((.((..(....)..))))))))).)))))((((((....)))))).))))))... (-28.52 = -28.15 + -0.37)

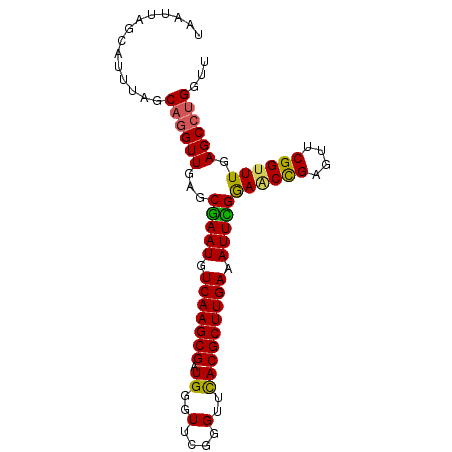
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:20 2006