
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,245,993 – 3,246,100 |
| Length | 107 |
| Max. P | 0.989734 |

| Location | 3,245,993 – 3,246,100 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -20.77 |
| Energy contribution | -19.90 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3245993 107 + 27905053 -------------GUACCAUCUACGUGAUGCUAUAUGCCAUUGUUAUCGGAGAGCACUUGGUAGCUUUCAAUAACUAUGAUGAGAGUUCUUAUAAGGUAUACCGUGUAGAAAGUAUAUAU -------------((((..(((((((((((((......(((.(((((..((((((........)))))).))))).)))(((((....)))))..)))))..))))))))..)))).... ( -26.90) >DroSim_CAF1 64 120 + 1 AAACAAUGCAUUGAUACCGUCGAUGUGAUGCGAUAUGCCAUUGUCACUGGAGAGCACUUGAUUGCUUUCAAAAACUACGAUGAGAGUCCUUAUAAGGUAUACCUCGUCGAAAGUAUACAU .......((((((((...)))))))).((((((((......))))....(((((((......)))))))........(((((((.(((((....)))...)))))))))...)))).... ( -31.20) >DroYak_CAF1 15803 120 + 1 AAACAAUGCAUUGAUACUGUCGAUGUGAUGCGAUAUGCCAUUAUCACUGGAGAGCACUCGAUUGCUUUUAAUGACUAUGACGAGAGUCCCUAUAAGGUAUACCUUAUAGGAAGCAUAUAU ...((.(((((((((...))))))))).))..((((((.....(((...(((((((......)))))))..)))....(((....)))(((((((((....)))))))))..)))))).. ( -37.50) >consensus AAACAAUGCAUUGAUACCGUCGAUGUGAUGCGAUAUGCCAUUGUCACUGGAGAGCACUUGAUUGCUUUCAAUAACUAUGAUGAGAGUCCUUAUAAGGUAUACCUUGUAGAAAGUAUAUAU .............((((((((...(((((((.....))....)))))..((((((........)))))).........)))).......((((((((....))))))))...)))).... (-20.77 = -19.90 + -0.87)

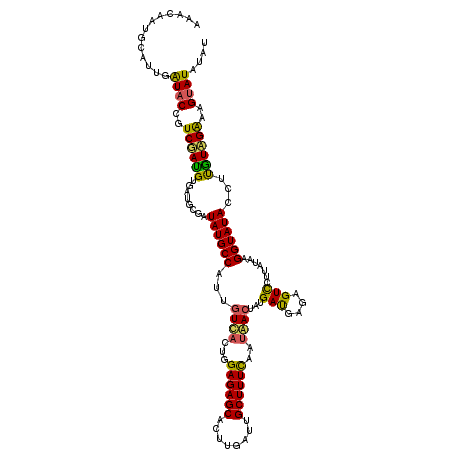
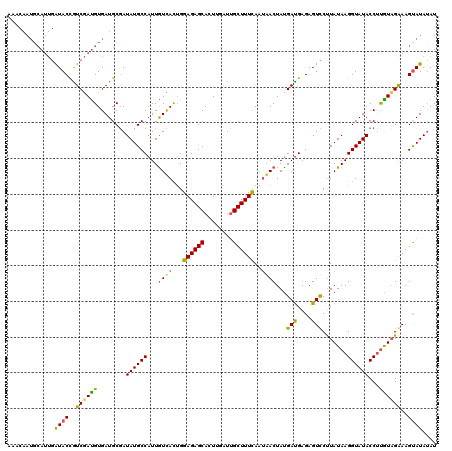
| Location | 3,245,993 – 3,246,100 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -18.31 |
| Energy contribution | -17.67 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3245993 107 - 27905053 AUAUAUACUUUCUACACGGUAUACCUUAUAAGAACUCUCAUCAUAGUUAUUGAAAGCUACCAAGUGCUCUCCGAUAACAAUGGCAUAUAGCAUCACGUAGAUGGUAC------------- .....((((.(((((..((....))......((........(((.(((((((..(((........)))...))))))).)))((.....)).))..))))).)))).------------- ( -22.60) >DroSim_CAF1 64 120 - 1 AUGUAUACUUUCGACGAGGUAUACCUUAUAAGGACUCUCAUCGUAGUUUUUGAAAGCAAUCAAGUGCUCUCCAGUGACAAUGGCAUAUCGCAUCACAUCGACGGUAUCAAUGCAUUGUUU ..(((((((((....)))))))))....((((((((........)))))))).(((((((((.(((((.((..((((.....((.....)).))))...)).)))))...)).))))))) ( -23.30) >DroYak_CAF1 15803 120 - 1 AUAUAUGCUUCCUAUAAGGUAUACCUUAUAGGGACUCUCGUCAUAGUCAUUAAAAGCAAUCGAGUGCUCUCCAGUGAUAAUGGCAUAUCGCAUCACAUCGACAGUAUCAAUGCAUUGUUU ....(((((((((((((((....))))))))))).....(((((.((((((...((((......))))....)))))).))))).....))))......((((((........)))))). ( -33.90) >consensus AUAUAUACUUUCUACAAGGUAUACCUUAUAAGGACUCUCAUCAUAGUUAUUGAAAGCAAUCAAGUGCUCUCCAGUGACAAUGGCAUAUCGCAUCACAUCGACGGUAUCAAUGCAUUGUUU .....((((.(((((((((....))))....((........(((.(((((((..(((........)))...))))))).)))((.....)).))..))))).)))).............. (-18.31 = -17.67 + -0.64)

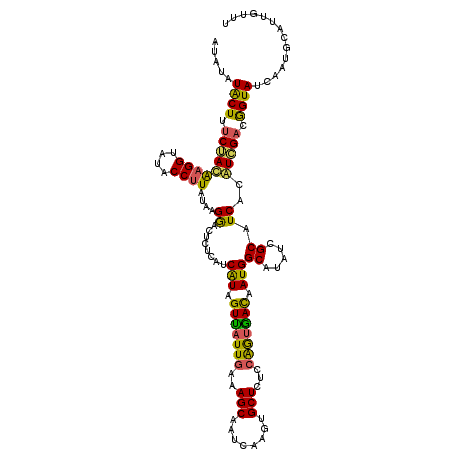
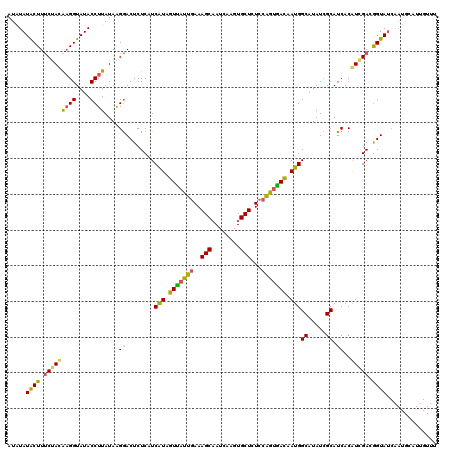
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:11 2006