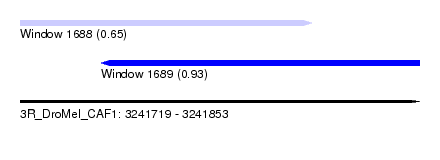
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,241,719 – 3,241,853 |
| Length | 134 |
| Max. P | 0.928089 |

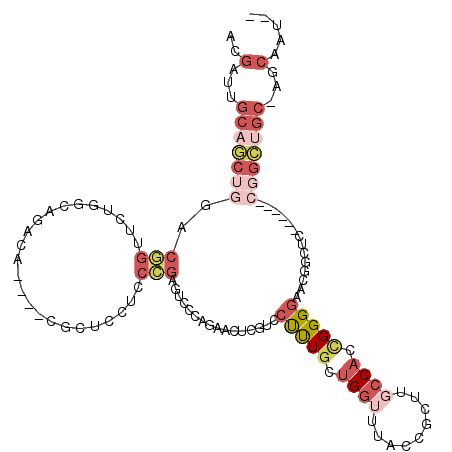
| Location | 3,241,719 – 3,241,817 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 68.80 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -10.67 |
| Energy contribution | -14.45 |
| Covariance contribution | 3.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3241719 98 + 27905053 -AAAAGGAGAGCAAACUUUAA-GUUUGCAAUUGCU-GCAGCCG------GAGCAGUUCCCCGGUGGCAAGCGGUAAACCAGCAAAGGACGAGUUCUGGGACUCGGGA -....((...((((((.....-)))))).((((((-((.((((------(.........))))).)).))))))...)).........((((((....))))))... ( -34.50) >DroVir_CAF1 10544 82 + 1 CAGAUCUAAAGCAAACUUUAAACUUUGCAAAUGCUAGCAAC----------GCUGCU-CCCUAAAGAAAAUGGUAAGCCAAAGGGGUGU------GUGU-------- ..........(((((........)))))........(((.(----------((.(((-(((....)....(((....)))..)))))))------))))-------- ( -14.00) >DroSec_CAF1 8785 98 + 1 -AAAAGGAGAGCAAACCUUAA-GUUUGCAAUUGCU-GCAGCCG------GAGCCGUUCCCCGGUGGCAAGCGGUAAACCAGCAAAGGACGAGUUCUGGGACUCGGGA -....((...((((((.....-)))))).((((((-((.((((------(.........))))).)).))))))...)).........((((((....))))))... ( -33.80) >DroSim_CAF1 9061 98 + 1 -AAAAGGAGAGCAAACUUUAA-GUUUGCAAUUGCU-GCAGCCG------GAGCCGUUCCCCGGUGGCAAGCGGUAAACCAGCAAAGGACGAGUUCUGGGACUCGGGA -....((...((((((.....-)))))).((((((-((.((((------(.........))))).)).))))))...)).........((((((....))))))... ( -34.50) >DroEre_CAF1 8968 104 + 1 -AAAAGCAGAGCAAACUUUAA-GUUUGCAAUUGCU-GCAGCCGGAGCCGGAGCCGUUCCCCGGUGGAAAGCGGUAAACCAGCAAAGGAUGAGUUCUGGGACUCGGGA -....((((.((((((.....-)))))).....))-))..((.(((((((((((((((((....))...((((....)).))...))))).)))))))..))).)). ( -37.10) >DroMoj_CAF1 11190 83 + 1 CAGAUCCAAGAGAAACUUUAAAGUUUGCAAGCGC-AGCUAC-----------------CCCAAAGGCCAGCGGUAAACCAGCAGGGGGU------UGGGGAGCUGCU ............(((((....)))))......((-((((.(-----------------(((((...((.((((....)).)).))...)------))))))))))). ( -29.70) >consensus _AAAAGGAGAGCAAACUUUAA_GUUUGCAAUUGCU_GCAGCCG______GAGCCGUUCCCCGGUGGCAAGCGGUAAACCAGCAAAGGACGAGUUCUGGGACUCGGGA ..........((((((......))))))....(((.((.((((.................)))).)).)))((....)).........((((((....))))))... (-10.67 = -14.45 + 3.78)

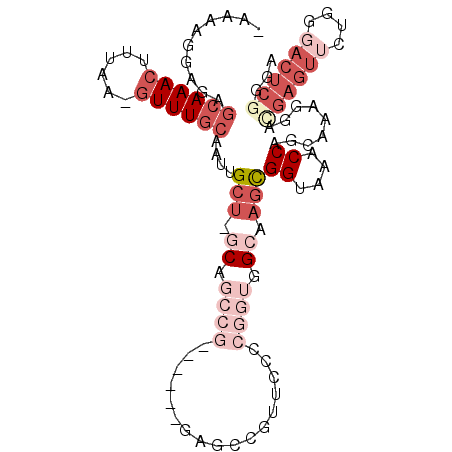
| Location | 3,241,746 – 3,241,853 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.21 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -8.87 |
| Energy contribution | -11.29 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.22 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3241746 107 - 27905053 ACGAUUGCAGCUGGACGGUUUUGGCAGACA----CGCUCCUCCCGAGUCCCAGAACUCGUCCUUUGCUGGUUUACCGCUUGCCACCGGGGAACUGCUC------CGGCUGC-AGCAAU-- .......((((.((((((((((((..(((.----((.......)).)))))))))).)))))...)))).......(((.((..((((((.....)))------)))..))-)))...-- ( -42.90) >DroSec_CAF1 8812 107 - 1 ACGAUUGCAGCUGGACGGUUCUGGCAGACA----CGCUCCUCCCGAGUCCCAGAACUCGUCCUUUGCUGGUUUACCGCUUGCCACCGGGGAACGGCUC------CGGCUGC-AGCAAU-- .......((((.((((((((((((..(((.----((.......)).)))))))))).)))))...)))).......(((.((..((((((.....)))------)))..))-)))...-- ( -45.50) >DroSim_CAF1 9088 107 - 1 ACGAUUGCAGCUGGACGGUUCUGGCAGACA----CGCUCCUCCCGAGUCCCAGAACUCGUCCUUUGCUGGUUUACCGCUUGCCACCGGGGAACGGCUC------CGGCUGC-AGCAAU-- .......((((.((((((((((((..(((.----((.......)).)))))))))).)))))...)))).......(((.((..((((((.....)))------)))..))-)))...-- ( -45.50) >DroEre_CAF1 8995 107 - 1 ACGAUUGCAGCUGGACGGGUCUGGG----------UCUCCUCCCGAGUCCCAGAACUCAUCCUUUGCUGGUUUACCGCUUUCCACCGGGGAACGGCUCCGGCUCCGGCUGC-AGCAAU-- ..(.(((((((((((.(((((((((----------.(((.....))).))))).)))).......(((((....(((.(((((....))))))))..))))))))))))))-)))...-- ( -52.80) >DroMoj_CAF1 11219 90 - 1 ACGAUUGCAACUGAACUGG----GCAGCAACAAAGCCACCAGCAGCUCCCCA------ACCCCCUGCUGGUUUACCGCUGGCCUUUGGG-----------------GUAGCU-GCGCU-- ......((........(((----...((......))..)))(((((((((((------(...((.((.((....)))).))...)))))-----------------).))))-)))).-- ( -34.40) >DroPer_CAF1 8326 86 - 1 CCAAUUGCAGCUGGACCUCCC-------CA----CCCUCCUGCUCGGCCC-------CCCUCUCUCCUGGUUUACCACUUUACAUUGGGGAACUGCU----------------GCUGUUG .((((.(((((.(....((((-------((----...........((...-------.)).......(((....)))........)))))).).)))----------------)).)))) ( -23.00) >consensus ACGAUUGCAGCUGGACGGUUCUGGCAGACA____CGCUCCUCCCGAGUCCCAGAACUCGUCCUUUGCUGGUUUACCGCUUGCCACCGGGGAACGGCUC______CGGCUGC_AGCAAU__ ..(...(((((((..(((........................)))................(((((.((((.........)))).)))))..............)))))))...)..... ( -8.87 = -11.29 + 2.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:08 2006