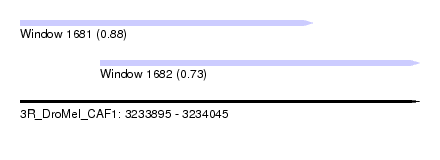
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,233,895 – 3,234,045 |
| Length | 150 |
| Max. P | 0.882425 |

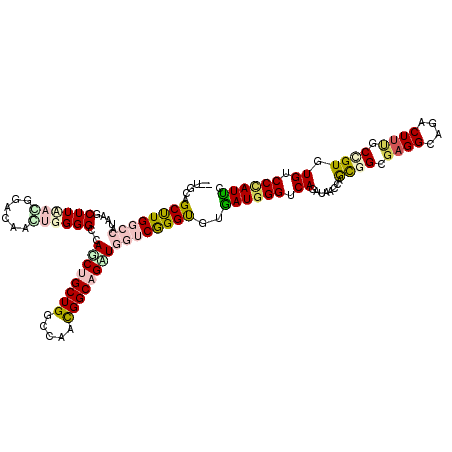
| Location | 3,233,895 – 3,234,005 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3233895 110 + 27905053 -------U--UAAUCAGAAUAUUAUCCCAU-CUUUUUGCCAGUGCAGCUUGGCCAUCAGCUUAACGGAUAAGUGGGGAGAGCUGCUGGACAAUGGCAGUUGGUCAGGUGUGAUGGGUCAG -------.--......((.(((((..((..-((..((((((..(((((((..((....(((((.....))))).))..))))))).......))))))..))...))..)))))..)).. ( -34.70) >DroVir_CAF1 402 89 + 1 ---------------------UUACUACAA----------CUUGCAGCUUGGAAGUGCGCUUUCGCGAAACUUGGGGGCAGCUGCUGCCAAAUGGCAGCUGGACGGGUGUAAUGGGCCAC ---------------------....((((.----------(((((.((((..((((.(((....)))..))))..))))....((((((....))))))..).))))))))......... ( -27.50) >DroSec_CAF1 1203 111 + 1 -------U--UAAUAAUAAUAUUUUCCCACCCUUUUUGCCAGUGCAGCUUGGCCAUCAGCUUAACGGAUAAGUGGGGCGAGCUGCUGGAGAAUGGCAGUUGGUCAGGUGUGAUGGGUCAG -------.--..............((.((((((..((((((..((((((((.((....(((((.....))))).)).)))))))).......))))))..))...)))).))........ ( -36.50) >DroSim_CAF1 1204 111 + 1 -------U--UAAUGAUAAUAUUUUCCCACCCUUUUUGCCAGUGCAGCUUGGCCAUCAGCUUAACGGAUAAGUGGGGCGAGCUGCUGGAGAAUGGCAGUUGGUCAGGUGUGAUGGGUCAG -------.--...((((.......((.((((((..((((((..((((((((.((....(((((.....))))).)).)))))))).......))))))..))...)))).))...)))). ( -38.60) >DroEre_CAF1 1261 115 + 1 GGAAUGAUUAUGAUCAGAAUAU----CCCC-CCUUUUUCCAGUGCAGCUUGGCCAUCAGCCUAACGGACAAGUGGGGCGAGCUGCUGGAGAACGGCAGUUGGUCAGGUGUGAUGGGUCAG (((.((((....)))).....)----))..-......((((..(((.(((((((....((((.((......)).)))).((((((((.....))))))))))))))))))..)))).... ( -39.90) >DroYak_CAF1 1282 110 + 1 -------U--UAAUCAGAAUAUUACACCAC-CUUUUUCCCAGUGCAGCUUGGCCAUAAGCUUAACGGAUAAGUGGGGCGAACUGCUGGAGAAUGGCAGUUGGUCAGGUGUGAUGGGUCAG -------.--......((.(((((((((.(-(.(((..((.((..((((((....))))))..))))..))).))(((.((((((((.....)))))))).))).)))))))))..)).. ( -39.10) >consensus _______U__UAAUCAGAAUAUUAUCCCAC_CUUUUUGCCAGUGCAGCUUGGCCAUCAGCUUAACGGAUAAGUGGGGCGAGCUGCUGGAGAAUGGCAGUUGGUCAGGUGUGAUGGGUCAG ......................................(((.((((.(((((((....((((.((......)).)))).((((((((.....))))))))))))))))))).)))..... (-26.19 = -26.58 + 0.39)

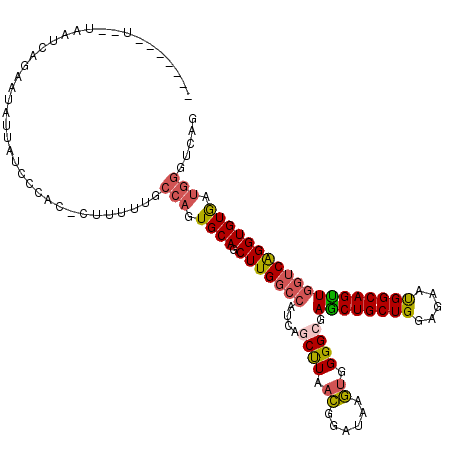
| Location | 3,233,925 – 3,234,045 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -48.40 |
| Consensus MFE | -28.14 |
| Energy contribution | -29.23 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3233925 120 + 27905053 AGUGCAGCUUGGCCAUCAGCUUAACGGAUAAGUGGGGAGAGCUGCUGGACAAUGGCAGUUGGUCAGGUGUGAUGGGUCAGGUGACGAGUCGCGAGGCGGACUUUGCCGUGUGUCCCAUUC ...(((((((..((....(((((.....))))).))..))))))).((((((((((((((.(((...((((((..(((....)))..)))))).))).)))..)))))).)))))..... ( -49.50) >DroVir_CAF1 411 120 + 1 CUUGCAGCUUGGAAGUGCGCUUUCGCGAAACUUGGGGGCAGCUGCUGCCAAAUGGCAGCUGGACGGGUGUAAUGGGCCACGUUACAAGCGGCGAGGUACACUUUGCUGUAUGCCCCAUUA ......((..(((((....)))))))........((((((...((((((....))))))........(((((((.....))))))).(((((((((....))))))))).)))))).... ( -47.50) >DroPse_CAF1 990 117 + 1 ---UCAGCCUGGCCAUAAGCUUCACGGACAACUGGGGCCAGCAGCUGGCCAACGGCAGAUGGUCGGGUGUGAUGGGUCAGAUAACCAACGGGGAGGCCGACUUUGCCGUGUGUCCCAUUC ---...((((((((((..(((...(((....))).((((((...))))))...)))..))))))))))..((((((.((........((((.((((....)))).)))).)).)))))). ( -47.50) >DroYak_CAF1 1312 120 + 1 AGUGCAGCUUGGCCAUAAGCUUAACGGAUAAGUGGGGCGAACUGCUGGAGAAUGGCAGUUGGUCAGGUGUGAUGGGUCAGGUGACAAGUCGCGCGGCGGACUUGGCCGUGUGUCCUAUUC ...((((.((.(((....(((((.....)))))..))).)))))).(((.((((((((((.(((..(((((((..(((....)))..)))))))))).))))..))))).).)))..... ( -46.60) >DroMoj_CAF1 421 120 + 1 GCUACAGCUUGCAGGUGCUCUUUCGCGACACCUGGGGUCAGCUGCUGCCCAACGGCAGCUGGUCGGGCGUGAUUGGCCACAUAGUCAGCGGCGAGGCAGACUUCUCCGUGUGCCCCAUCC ((....))...((((((.((......)))))))).(..(((((((((.....)))))))))..)(((((((((((......))))))((((.((((....)))).)))).)))))..... ( -51.80) >DroPer_CAF1 980 117 + 1 ---UCAGCCUGGCCAUAAGCUUCACGGACAACUGGGGCCAGCAGCUGGCCAACGGCAGAUGGUCGGGUGUGAUGGGUCAGAUAACCAACGGGGAGGCCGACUUUGCCGUGUGUCCCAUUC ---...((((((((((..(((...(((....))).((((((...))))))...)))..))))))))))..((((((.((........((((.((((....)))).)))).)).)))))). ( -47.50) >consensus __UGCAGCUUGGCCAUAAGCUUAACGGACAACUGGGGCCAGCUGCUGGCCAACGGCAGAUGGUCGGGUGUGAUGGGUCAGAUAACCAGCGGCGAGGCAGACUUUGCCGUGUGUCCCAUUC ......((((((((.....((((((......))))))..((((((((.....))))))))))))))))..((((((.((........((((.((((....)))).)))).)).)))))). (-28.14 = -29.23 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:01 2006