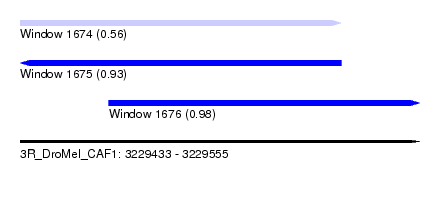
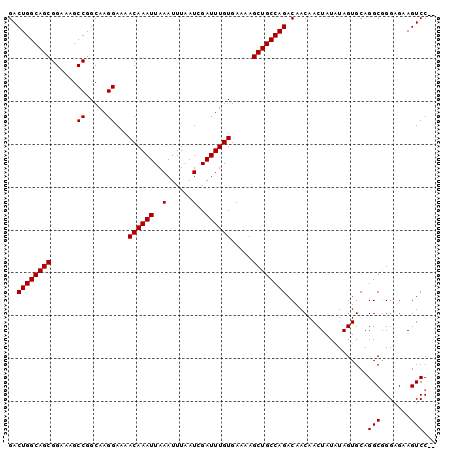
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,229,433 – 3,229,555 |
| Length | 122 |
| Max. P | 0.979558 |

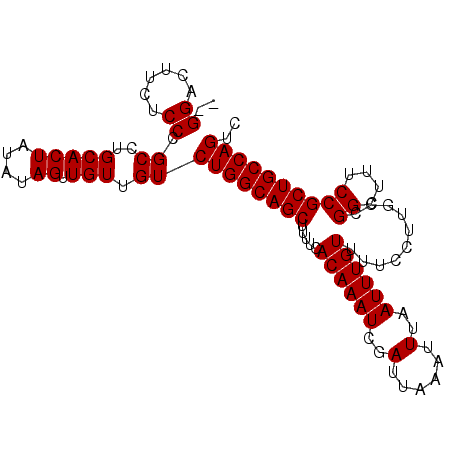
| Location | 3,229,433 – 3,229,531 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3229433 98 + 27905053 --GGACUUCUCCCGCCUGCACUAUAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCCGCUGCCAGUC --(((....))).((..(((((....)).))).))((((((((.....((((((..(......)..))))))..........((....)))))))))).. ( -21.00) >DroSec_CAF1 54006 98 + 1 --GGACUUCUCCCGCCUGCACUAUAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCCGCUGCCAGUC --(((....))).((..(((((....)).))).))((((((((.....((((((..(......)..))))))..........((....)))))))))).. ( -21.00) >DroSim_CAF1 55426 98 + 1 --GGACUUCUCCCGCCUGCACUAUAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCCGCUGCCAGUC --(((....))).((..(((((....)).))).))((((((((.....((((((..(......)..))))))..........((....)))))))))).. ( -21.00) >DroEre_CAF1 48409 100 + 1 CCGGACUUCGCCCGCCUGCACUACAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCCGCUGCCAGUC .(((.......))).....((.(((....))).))((((((((.....((((((..(......)..))))))..........((....)))))))))).. ( -22.40) >DroYak_CAF1 39875 98 + 1 --GGACUUCGCCUGCCUGCACUAUAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCCGCUGCCAGUC --.((((.....((....))......)))).....((((((((.....((((((..(......)..))))))..........((....)))))))))).. ( -20.90) >consensus __GGACUUCUCCCGCCUGCACUAUAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCCGCUGCCAGUC ..((......)).((..(((((....)).))).))((((((((.....((((((..(......)..))))))..........((....)))))))))).. (-20.50 = -20.50 + 0.00)

| Location | 3,229,433 – 3,229,531 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.86 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3229433 98 - 27905053 GACUGGCAGCGGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUAUAGUGCAGGCGGGAGAAGUCC-- ..((((((((......((.....))...((((((..(......)..)))))).....))))))))...................(((.......))).-- ( -23.00) >DroSec_CAF1 54006 98 - 1 GACUGGCAGCGGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUAUAGUGCAGGCGGGAGAAGUCC-- ..((((((((......((.....))...((((((..(......)..)))))).....))))))))...................(((.......))).-- ( -23.00) >DroSim_CAF1 55426 98 - 1 GACUGGCAGCGGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUAUAGUGCAGGCGGGAGAAGUCC-- ..((((((((......((.....))...((((((..(......)..)))))).....))))))))...................(((.......))).-- ( -23.00) >DroEre_CAF1 48409 100 - 1 GACUGGCAGCGGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUGUAGUGCAGGCGGGCGAAGUCCGG ..((((((((......((.....))...((((((..(......)..)))))).....))))))))((.(((....))).)).....(((((...))))). ( -27.90) >DroYak_CAF1 39875 98 - 1 GACUGGCAGCGGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUAUAGUGCAGGCAGGCGAAGUCC-- ..((((((((......((.....))...((((((..(......)..)))))).....))))))))...............(((......)))......-- ( -23.00) >consensus GACUGGCAGCGGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUAUAGUGCAGGCGGGAGAAGUCC__ ..((((((((......((.....))...((((((..(......)..)))))).....))))))))...................(((.......)))... (-22.86 = -22.86 + -0.00)

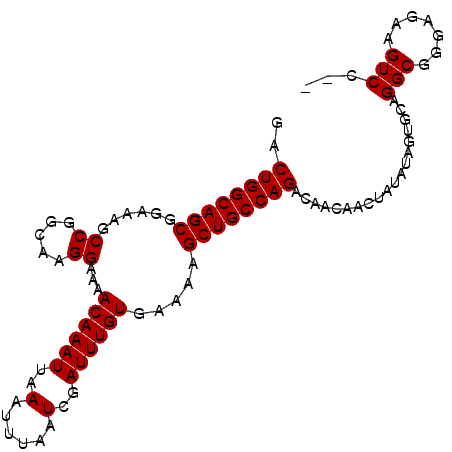
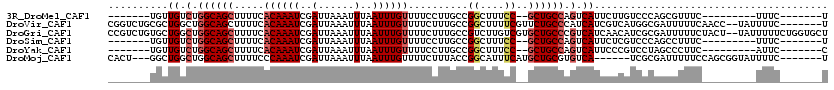
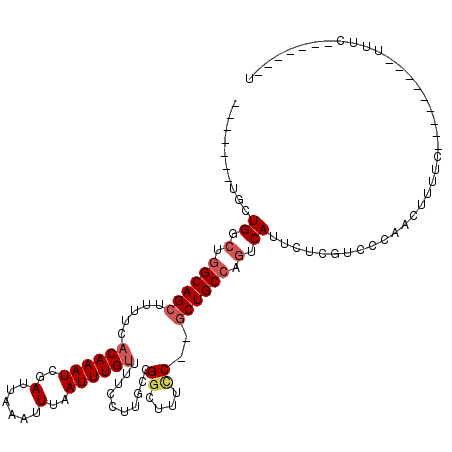
| Location | 3,229,460 – 3,229,555 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.81 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -11.37 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3229460 95 + 27905053 -------UGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCC--GCUGCCAGUCAUUCUUGUCCCAGCGUUUC---------UUUC-------U -------...((.((((((((.....((((((..(......)..))))))..........((....))--)))))))).))..................---------....-------. ( -22.00) >DroVir_CAF1 61718 111 + 1 CGGUCUGCGCUGGCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCUUUGCCGGCUUUUCGUUCUGCCCAUCAUCGUCAUGGCGAUUUUUCAACC--UAUUUUC-------U .(((..(((..(((((((((......((((((..(......)..)))))).....)))))))))...)))...)))....(((((....))))).........--.......-------. ( -28.20) >DroGri_CAF1 52567 118 + 1 CCGUCUGUGCUGGCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCUUUGCCGUCUUGUCGUGCUGCCCGUCAUCAACAUCGCGAUUUUUCUACU--UAUUUUUCUGGUGCU .(((.(((..((((.((((((.....((((((..(......)..)))))).........((......)).)))))).))))...)))..)))...........--............... ( -23.90) >DroSim_CAF1 55453 95 + 1 -------UGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCC--GCUGCCAGUCAUUCUCGUCCCAGCCUUUC---------UUUC-------U -------...((.((((((((.....((((((..(......)..))))))..........((....))--)))))))).))..................---------....-------. ( -22.00) >DroYak_CAF1 39902 95 + 1 -------UGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCC--GCUGCCAGUCAUUCCCGUCCUAGCCCUUC---------AUUC-------C -------...((.((((((((.....((((((..(......)..))))))..........((....))--)))))))).))..................---------....-------. ( -22.00) >DroMoj_CAF1 68806 104 + 1 CACU---GGCUGGCUGGCAGCUUUUCCCAAAUCGAUUAAAUUUAAUUUGUUUUCUUUACCGGCAUUUCAUGCUGCGUGUCA------UCGCGAUUUUUCCAGCGGUAUUUUC-------U .(((---(.((((...(((((......(((((..(......)..)))))...........((....))..)))))(((...------.))).......))))))))......-------. ( -20.50) >consensus _______UGCUGGCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCC__GCUGCCAGUCAUUCUCGUCCCAACUUUUC_________UUUC_______U ..........((.(.((((((.....((((((..(......)..))))))..........((....))..)))))).).))....................................... (-11.37 = -12.03 + 0.67)
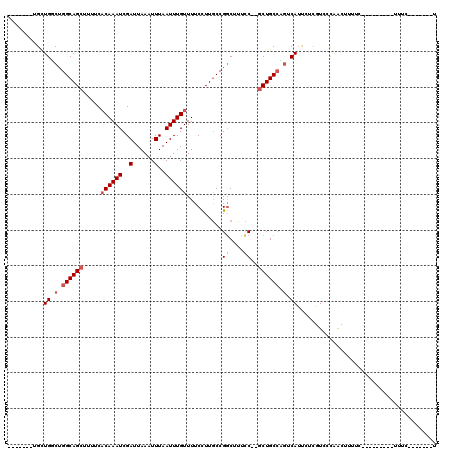
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:55 2006