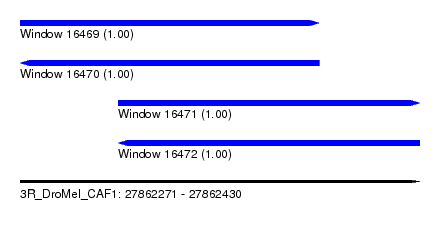
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,862,271 – 27,862,430 |
| Length | 159 |
| Max. P | 0.999722 |

| Location | 27,862,271 – 27,862,390 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -32.52 |
| Energy contribution | -32.28 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27862271 119 + 27905053 UUUGAAUUAUUAAUUUUUAAACUUUU-CGCAACACAAAAAGAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGC .........................(-((((.(((........(((((((.(((((........))))).)))))))......((((.....))))))).)))))((..((...))..)) ( -35.70) >DroSec_CAF1 171521 119 + 1 UUUGAAUUAUUAAUUUUUAAACUUUU-CGCAACACAAAAAGAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGC .........................(-((((.(((........(((((((.(((((........))))).)))))))......((((.....))))))).)))))((..((...))..)) ( -35.70) >DroSim_CAF1 173738 119 + 1 UUUGAAUUAUUAAUUUUUAAACUUUU-CGCAACACAAAAAGAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGC .........................(-((((.(((........(((((((.(((((........))))).)))))))......((((.....))))))).)))))((..((...))..)) ( -35.70) >DroEre_CAF1 201597 120 + 1 UUUGAAUUAUUAAUUUUUAAGCUUUUCCGCAGCACAAAAAGAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUAAAGUAAUAGGGUGAUGCGAGGGUGGUACCCUUGC ............(((((...(((((...((..(((........(((((((.(((((........))))).))))))))))..)).)))))...)))))...(((((((.....))))))) ( -39.90) >DroYak_CAF1 175373 120 + 1 UUUGAAUUAUUAAUUUUUAAGCUUUUCCGCAGCACAAAAAGAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUAAAGUAAUAGGGUGAUGCGAGGGUGGUACCCUUGC ............(((((...(((((...((..(((........(((((((.(((((........))))).))))))))))..)).)))))...)))))...(((((((.....))))))) ( -39.90) >consensus UUUGAAUUAUUAAUUUUUAAACUUUU_CGCAACACAAAAAGAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGC ....................((((....((..(((........(((((((.(((((........))))).))))))))))..))..))))...((((((...((....)).))))))... (-32.52 = -32.28 + -0.24)

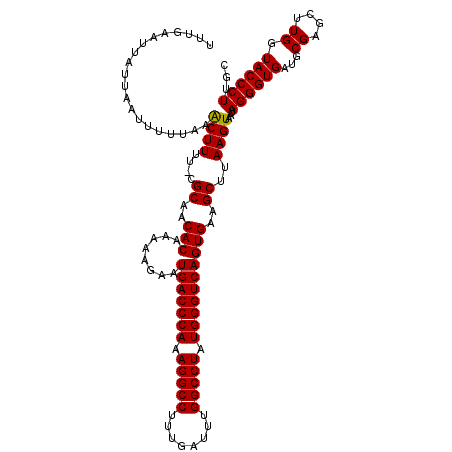
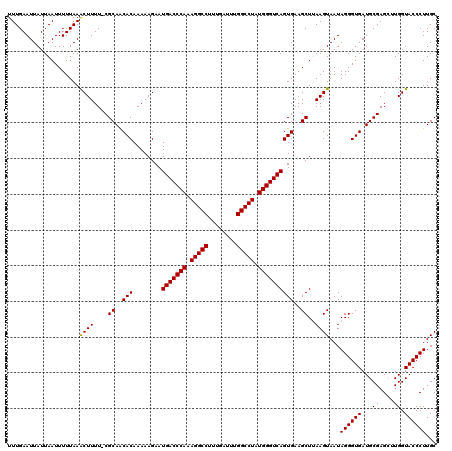
| Location | 27,862,271 – 27,862,390 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.06 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27862271 119 - 27905053 GCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUCUUUUUGUGUUGCG-AAAAGUUUAAAAAUUAAUAAUUCAAA ...(((((..............))))).....((((((.....(((((((.(((((........))))).)))))))...((((((.....))-))))))))))................ ( -32.74) >DroSec_CAF1 171521 119 - 1 GCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUCUUUUUGUGUUGCG-AAAAGUUUAAAAAUUAAUAAUUCAAA ...(((((..............))))).....((((((.....(((((((.(((((........))))).)))))))...((((((.....))-))))))))))................ ( -32.74) >DroSim_CAF1 173738 119 - 1 GCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUCUUUUUGUGUUGCG-AAAAGUUUAAAAAUUAAUAAUUCAAA ...(((((..............))))).....((((((.....(((((((.(((((........))))).)))))))...((((((.....))-))))))))))................ ( -32.74) >DroEre_CAF1 201597 120 - 1 GCAAGGGUACCACCCUCGCAUCACCCUAUUACUUUAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUCUUUUUGUGCUGCGGAAAAGCUUAAAAAUUAAUAAUUCAAA ((.((((.....))))((((.(((.(((......)))......(((((((.(((((........))))).)))))))........))).)))).....)).................... ( -37.40) >DroYak_CAF1 175373 120 - 1 GCAAGGGUACCACCCUCGCAUCACCCUAUUACUUUAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUCUUUUUGUGCUGCGGAAAAGCUUAAAAAUUAAUAAUUCAAA ((.((((.....))))((((.(((.(((......)))......(((((((.(((((........))))).)))))))........))).)))).....)).................... ( -37.40) >consensus GCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUCUUUUUGUGUUGCG_AAAAGUUUAAAAAUUAAUAAUUCAAA ((.(((((..............))))).........((..((((((((((.(((((........))))).)))))))........)))..))......)).................... (-31.30 = -31.06 + -0.24)

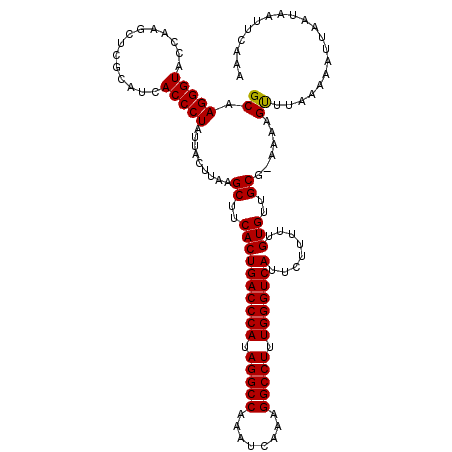
| Location | 27,862,310 – 27,862,430 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -44.32 |
| Consensus MFE | -40.90 |
| Energy contribution | -40.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
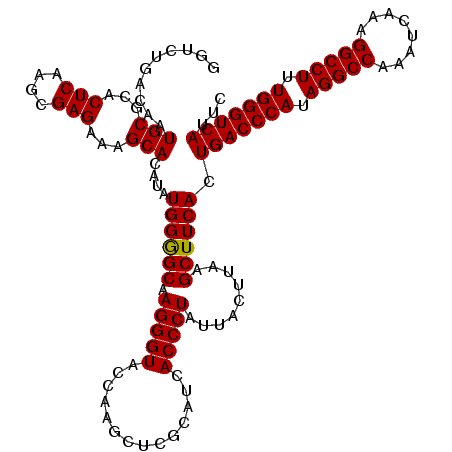
>3R_DroMel_CAF1 27862310 120 + 27905053 GAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGCCCCAUAUGUGCUUUCUCGCUUGAGUGCGCAUUGUCAGACC ...(((((((.(((((........))))).)))))))(((..(((((((..(.(((((.(((.(.((..((...))..)).))))....))))).)..))))))).)))........... ( -41.70) >DroSec_CAF1 171560 120 + 1 GAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGCCCCAUAUGUGCUUUCUCGCUUGAGUGCGCAUUGUCAGACC ...(((((((.(((((........))))).)))))))(((..(((((((..(.(((((.(((.(.((..((...))..)).))))....))))).)..))))))).)))........... ( -41.70) >DroSim_CAF1 173777 120 + 1 GAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGCCCCAUAUGUGCUUUCUCGCUUGAGUGCGCAUUGUCAGACC ...(((((((.(((((........))))).)))))))(((..(((((((..(.(((((.(((.(.((..((...))..)).))))....))))).)..))))))).)))........... ( -41.70) >DroEre_CAF1 201637 119 + 1 GAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUAAAGUAAUAGGGUGAUGCGAGGGUGGUACCCUUGCCCCAUAUGUGCU-UCUCGCUUGAGUGCGCAUUGUCAGACC ...(((((((.(((((........))))).))))))).(((.............(((....(((((((.....))))))))))..((((((.-.(((....))).))))))..))).... ( -48.80) >DroYak_CAF1 175413 120 + 1 GAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUAAAGUAAUAGGGUGAUGCGAGGGUGGUACCCUUGCUCCAUAUGUGCUUUCUCGCUUGAGUGCGCAUUGUCAGACC ...(((((((.(((((........))))).))))))).................((.(((((((((((.....))))))).....((((((...(((....))).)))))).))))..)) ( -47.70) >consensus GAAUGACCCAAAGGCCUUUGAUUUGGCCUAUGGGUCAGUGAAGCUUAAGUAAUAGGGUGAUGCGAGCUUGGUACCCUUGCCCCAUAUGUGCUUUCUCGCUUGAGUGCGCAUUGUCAGACC ...(((((((.(((((........))))).)))))))..................((..(((((((...(((((...((...))...)))))..)))((......))))))..))..... (-40.90 = -40.90 + -0.00)



| Location | 27,862,310 – 27,862,430 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -35.76 |
| Energy contribution | -35.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27862310 120 - 27905053 GGUCUGACAAUGCGCACUCAAGCGAGAAAGCACAUAUGGGGCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUC ............(((......))).(((.((....((((((...((((.....))))......)))))).......)))))..(((((((.(((((........))))).)))))))... ( -37.10) >DroSec_CAF1 171560 120 - 1 GGUCUGACAAUGCGCACUCAAGCGAGAAAGCACAUAUGGGGCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUC ............(((......))).(((.((....((((((...((((.....))))......)))))).......)))))..(((((((.(((((........))))).)))))))... ( -37.10) >DroSim_CAF1 173777 120 - 1 GGUCUGACAAUGCGCACUCAAGCGAGAAAGCACAUAUGGGGCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUC ............(((......))).(((.((....((((((...((((.....))))......)))))).......)))))..(((((((.(((((........))))).)))))))... ( -37.10) >DroEre_CAF1 201637 119 - 1 GGUCUGACAAUGCGCACUCAAGCGAGA-AGCACAUAUGGGGCAAGGGUACCACCCUCGCAUCACCCUAUUACUUUAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUC ............(((......))).((-(((....(((((((.((((.....)))).)).....))))).......)))))..(((((((.(((((........))))).)))))))... ( -42.70) >DroYak_CAF1 175413 120 - 1 GGUCUGACAAUGCGCACUCAAGCGAGAAAGCACAUAUGGAGCAAGGGUACCACCCUCGCAUCACCCUAUUACUUUAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUC (((....(((((.((.(((....)))...)).))).))((((.((((.....)))).)).)))))..................(((((((.(((((........))))).)))))))... ( -39.10) >consensus GGUCUGACAAUGCGCACUCAAGCGAGAAAGCACAUAUGGGGCAAGGGUACCAAGCUCGCAUCACCCUAUUACUUAAGCUUCACUGACCCAUAGGCCAAAUCAAAGGCCUUUGGGUCAUUC ..........(((...(((....)))...)))....((((((.(((((..............))))).........)))))).(((((((.(((((........))))).)))))))... (-35.76 = -35.60 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:38 2006