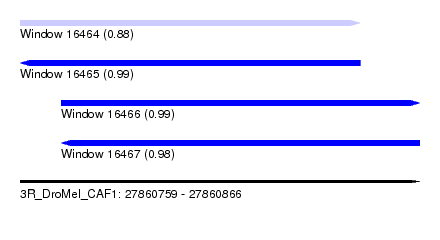
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,860,759 – 27,860,866 |
| Length | 107 |
| Max. P | 0.994882 |

| Location | 27,860,759 – 27,860,850 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -17.78 |
| Energy contribution | -17.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27860759 91 + 27905053 UUAAACAACUAAAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUAAAAGCACCUGUGCCUGCCGUAG ....((.(((......((((((.....))))))..........((((((..........)))))))))....(((....))).....)).. ( -17.30) >DroSec_CAF1 169983 91 + 1 UUAAACAACUAAAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAGCACCUGUGCCAGCCACAG ..............((((((((.....)))))((((((.....((((((..........))))))..)))))))))(((((.....))))) ( -21.90) >DroSim_CAF1 172208 91 + 1 UUAAACAACUAAAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAGCACCUGUGCCAGCCACAG ..............((((((((.....)))))((((((.....((((((..........))))))..)))))))))(((((.....))))) ( -21.90) >DroEre_CAF1 200049 91 + 1 UUAAACAACUAAAAGUGUCACAAUUGUUGUGACUUGUAACCAGGACCAAUUGUUUAACUUUGGUUAGCACAAACACCUGUGCCUGCUGAAG ((((((((.........(((((.....)))))((((....)))).....))))))))(((..((..(((((......)))))..))..))) ( -23.70) >DroYak_CAF1 173781 91 + 1 UUAAACAACUAAAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAACACCUGUGCCUUUCGGAG ........(..((((.((((((.....))))))..........((((((..........)))))).(((((......)))))))))..).. ( -20.50) >consensus UUAAACAACUAAAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAGCACCUGUGCCUGCCGCAG (((((((((.....))((((((.....)))))).................)))))))((.((((..(((((......)))))..)))).)) (-17.78 = -17.46 + -0.32)



| Location | 27,860,759 – 27,860,850 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 93.96 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27860759 91 - 27905053 CUACGGCAGGCACAGGUGCUUUUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUUUAGUUGUUUAA ....((((((((....))).....))).))....((((((((((((....(((.....((((((.....))))))))).)))))))))))) ( -21.10) >DroSec_CAF1 169983 91 - 1 CUGUGGCUGGCACAGGUGCUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUUUAGUUGUUUAA (((((.....)))))(((((((((((((((((..........))))))..)).)))))).)))((((((((.........))))))))... ( -25.40) >DroSim_CAF1 172208 91 - 1 CUGUGGCUGGCACAGGUGCUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUUUAGUUGUUUAA (((((.....)))))(((((((((((((((((..........))))))..)).)))))).)))((((((((.........))))))))... ( -25.40) >DroEre_CAF1 200049 91 - 1 CUUCAGCAGGCACAGGUGUUUGUGCUAACCAAAGUUAAACAAUUGGUCCUGGUUACAAGUCACAACAAUUGUGACACUUUUAGUUGUUUAA ........(((((((....)))))))........((((((((((((...((....)).((((((.....))))))....)))))))))))) ( -22.40) >DroYak_CAF1 173781 91 - 1 CUCCGAAAGGCACAGGUGUUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUUUAGUUGUUUAA ..((....)).((((.((.(((((((((((((..........))))))..)).)))))((((((.....)))))).....)).)))).... ( -21.80) >consensus CUGCGGCAGGCACAGGUGCUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUUUAGUUGUUUAA .....(((((((....)))))))(((......)))(((((((((((....(((.....((((((.....))))))))).))))))))))). (-19.76 = -19.84 + 0.08)

| Location | 27,860,770 – 27,860,866 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.06 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27860770 96 + 27905053 AAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUAAAAGCACCUGUGCCUGCCGUAGCAGAAUUGCAAAACAU .....((((((.....))))))((((((....((((((..........)))))).......(((....)))((((....))))..))))))..... ( -24.00) >DroSec_CAF1 169994 96 + 1 AAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAGCACCUGUGCCAGCCACAGCAGAAUUACAAAACAU ...((((((((.....)))))((((((.....((((((..........))))))..)))))))))(((((.....)))))................ ( -22.30) >DroSim_CAF1 172219 96 + 1 AAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAGCACCUGUGCCAGCCACAGCAGAAUUACAAAACAU ...((((((((.....)))))((((((.....((((((..........))))))..)))))))))(((((.....)))))................ ( -22.30) >DroEre_CAF1 200060 96 + 1 AAAGUGUCACAAUUGUUGUGACUUGUAACCAGGACCAAUUGUUUAACUUUGGUUAGCACAAACACCUGUGCCUGCUGAAGCAGAACUACAAAACAU .....((((((.....))))))(((((.....((((((..........)))))).(((((......)))))((((....))))...)))))..... ( -25.20) >DroYak_CAF1 173792 96 + 1 AAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAACACCUGUGCCUUUCGGAGCAGAACUACAAAACAU ..(((((((((.....))))))(((((.....((((((..........))))))..)))))....((((.((....)).)))).)))......... ( -23.50) >consensus AAAGUGUCACAAUUGUUGUGACUUGUAACUAGGACCAAUUGUUUAACUUUGGUCAGUACAAGCACCUGUGCCUGCCGCAGCAGAAUUACAAAACAU .....((((((.....))))))(((((.....((((((..........)))))).(((((......)))))((((....))))...)))))..... (-20.90 = -21.06 + 0.16)


| Location | 27,860,770 – 27,860,866 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -21.00 |
| Energy contribution | -20.44 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27860770 96 - 27905053 AUGUUUUGCAAUUCUGCUACGGCAGGCACAGGUGCUUUUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUU .(((...((....((((....))))(((....))).......((((((..........))))))...)).))).((((((.....))))))..... ( -25.00) >DroSec_CAF1 169994 96 - 1 AUGUUUUGUAAUUCUGCUGUGGCUGGCACAGGUGCUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUU ..((.(..(((((...(((((.....)))))(((((((((((((((((..........))))))..)).)))))).)))...)))))..).))... ( -25.30) >DroSim_CAF1 172219 96 - 1 AUGUUUUGUAAUUCUGCUGUGGCUGGCACAGGUGCUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUU ..((.(..(((((...(((((.....)))))(((((((((((((((((..........))))))..)).)))))).)))...)))))..).))... ( -25.30) >DroEre_CAF1 200060 96 - 1 AUGUUUUGUAGUUCUGCUUCAGCAGGCACAGGUGUUUGUGCUAACCAAAGUUAAACAAUUGGUCCUGGUUACAAGUCACAACAAUUGUGACACUUU .....((((((.(((((....)))))..((((...((((..((((....)))).)))).....)))).))))))((((((.....))))))..... ( -25.90) >DroYak_CAF1 173792 96 - 1 AUGUUUUGUAGUUCUGCUCCGAAAGGCACAGGUGUUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUU ..((.(..((((((((..((....))..)))(((((((((((((((((..........))))))..)).)))))).)))...)))))..).))... ( -24.60) >consensus AUGUUUUGUAAUUCUGCUGCGGCAGGCACAGGUGCUUGUACUGACCAAAGUUAAACAAUUGGUCCUAGUUACAAGUCACAACAAUUGUGACACUUU ..((.(..(((((.((((......))))...(((((((((((((((((..........))))))..)).)))))).)))...)))))..).))... (-21.00 = -20.44 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:34 2006