
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,859,725 – 27,859,885 |
| Length | 160 |
| Max. P | 0.998006 |

| Location | 27,859,725 – 27,859,845 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -24.76 |
| Energy contribution | -25.80 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27859725 120 - 27905053 CCAAAAAGGACCAGAGGACAAAGCCGAAAUGCCUGAGGCACCCACUGCUGCAGCUUGGCAUAAUAUUAAUUUAAAUUAGUCAAAAUGCCAGGCCUUUGGAAGGUCUUUUAGUAGAGAAGC ((.....))((.(((((((....(((((.(((....((((.....)))))))(((((((((...((((((....))))))....))))))))).)))))...))))))).))........ ( -32.60) >DroSec_CAF1 168963 120 - 1 CCAAAAAGGACCAGAGGACAAAGCCGAAAUGCCUGAGGCACCCACUGCUGCAGCUUGGCAUAAUAUUAAUUUAAAUUAGUCAAAAUGGCAGGCCUCUGGAAGGUCUUUCAGUAGAGAAGC ((.....)).(((((((.(...(((((..(((....((((.....)))))))..)))))...................(((.....))).).)))))))....((((......))))... ( -31.20) >DroSim_CAF1 171166 120 - 1 CCAAAAAGGACCAGAGGACAAAGCCGAAAUGCCUGAGGCACCCACUGCUGCAGCUUGGCAUAAUAUUAAUUUAAAUUAGUCAAAAUGGCAGGCCUCUGGAAGGUCUUUCAGUAGAGAAGC ((.....)).(((((((.(...(((((..(((....((((.....)))))))..)))))...................(((.....))).).)))))))....((((......))))... ( -31.20) >DroEre_CAF1 199075 120 - 1 CCAAAAAGGACCAGAGGACAAAGCCGAAAUGCCAGAGGCAGCCACUGCUGCAGCUUGGCAUAAUAUUAAUUUAAAUUAGACAAAAUGCCAGGCCUCUAGAAGGACUUUCAGUAGGAGAGU ((.....((......((......))......))(((((((((....))))).(((((((((....(((((....))))).....)))))))))))))....))((((((.....)))))) ( -35.30) >DroYak_CAF1 172729 120 - 1 CCAAAAAGGACCAGAGGACAAAGCCGAAAUGCCUGAGGCAGCCUCUGCUGCAGCUUCGCAUAAUAUUAAUUUAAAUUAGACAAAAUGCCAGGCCUCUAGAAGGUCUUUCAGUAGAAAAGC .....(((((((...((......)).....(((((..(((((....)))))......((((....(((((....))))).....)))))))))........)))))))............ ( -28.20) >consensus CCAAAAAGGACCAGAGGACAAAGCCGAAAUGCCUGAGGCACCCACUGCUGCAGCUUGGCAUAAUAUUAAUUUAAAUUAGUCAAAAUGCCAGGCCUCUGGAAGGUCUUUCAGUAGAGAAGC .....(((((((((((.....(((.(...((((...))))....).)))...(((((((((...((((((....))))))....)))))))))))))....)))))))............ (-24.76 = -25.80 + 1.04)


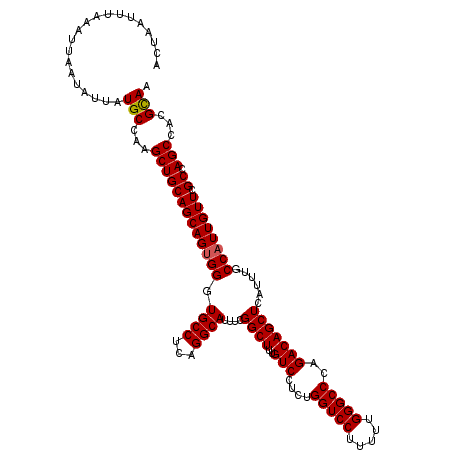
| Location | 27,859,765 – 27,859,885 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -33.76 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27859765 120 + 27905053 ACUAAUUUAAAUUAAUAUUAUGCCAAGCUGCAGCAGUGGGUGCCUCAGGCAUUUCGGCUUUGUCCUCUGGUCCUUUUUGGGCCCAGACAGCUCAUUUGCCAUUGUUCGCCAGCCACGCAA ....................(((...((((((((((((((((((...)))))...((((..(((....(((((.....)))))..)))))))......)))))))).).))))...))). ( -36.40) >DroSec_CAF1 169003 120 + 1 ACUAAUUUAAAUUAAUAUUAUGCCAAGCUGCAGCAGUGGGUGCCUCAGGCAUUUCGGCUUUGUCCUCUGGUCCUUUUUGGGCCCAGACAGCUCAUUUGCCAUUGUUCGCCAGCCACGCAA ....................(((...((((((((((((((((((...)))))...((((..(((....(((((.....)))))..)))))))......)))))))).).))))...))). ( -36.40) >DroSim_CAF1 171206 120 + 1 ACUAAUUUAAAUUAAUAUUAUGCCAAGCUGCAGCAGUGGGUGCCUCAGGCAUUUCGGCUUUGUCCUCUGGUCCUUUUUGGGCCCAGACAGCUCAUUUGCCAUUGUUCGCCAGCCACGCAA ....................(((...((((((((((((((((((...)))))...((((..(((....(((((.....)))))..)))))))......)))))))).).))))...))). ( -36.40) >DroEre_CAF1 199115 120 + 1 UCUAAUUUAAAUUAAUAUUAUGCCAAGCUGCAGCAGUGGCUGCCUCUGGCAUUUCGGCUUUGUCCUCUGGUCCUUUUUGGGCCCAGACAGCUCAUUGGCCAUUGUUCGCCAGCCACGCAA ....................(((...((((((((((((((((((...))).....((((..(((....(((((.....)))))..)))))))....)))))))))).).))))...))). ( -39.00) >DroYak_CAF1 172769 120 + 1 UCUAAUUUAAAUUAAUAUUAUGCGAAGCUGCAGCAGAGGCUGCCUCAGGCAUUUCGGCUUUGUCCUCUGGUCCUUUUUGGGCCCAGACAGCUCAUUGGCCAUUGUUCGCUAGCCACGUAA ....................((((..(((((((((((((.((((...))))))))((((..((..(((((.(((....))).)))))..)).....))))..)))).)).)))..)))). ( -36.30) >consensus ACUAAUUUAAAUUAAUAUUAUGCCAAGCUGCAGCAGUGGGUGCCUCAGGCAUUUCGGCUUUGUCCUCUGGUCCUUUUUGGGCCCAGACAGCUCAUUUGCCAUUGUUCGCCAGCCACGCAA ....................(((...(((((((((((((.((((...))))....((((..(((....(((((.....)))))..)))))))......)))))))).)).)))...))). (-33.76 = -33.80 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:31 2006