
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,766,020 – 27,766,160 |
| Length | 140 |
| Max. P | 0.873712 |

| Location | 27,766,020 – 27,766,134 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -21.16 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
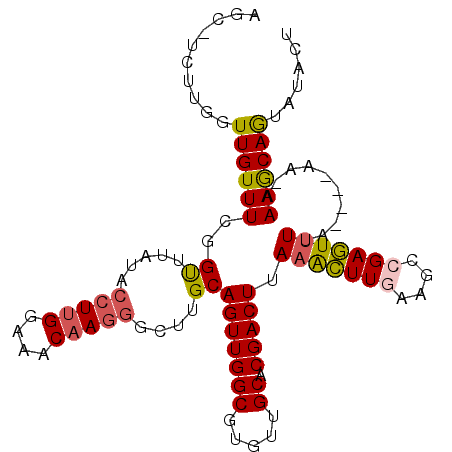
>3R_DroMel_CAF1 27766020 114 - 27905053 CUCGGAAACAAGGGCUUGCAGUUGGCGUGUUGCACGACUUAAACUUGUAGCCGAGUCUAA----AAAAAAACAGU--ACUUAAUUUUUAAUUAGCAACAUGUAAGUUAUAACAUAACUGU (((((..(((((...(((.(((((((.....)).)))))))).)))))..))))).....----......(((((--..........(((((.((.....)).))))).......))))) ( -24.23) >DroSec_CAF1 80415 115 - 1 CUUGGAAACAAGAGCUUGCAGUUGGCGUGUUGCACGACUUAAACUUGAAGCCGAGUUUUA----AA-AAAGCAGUAUACUUAAUUUGUAAUUAGCAACAUGUAAGUUAUAACAUAACUGU ((((....)))).....((((((((((((((((((..((((((((((....)))))))..----..-.)))..)).(((.......)))....)))))))))..((....)).))))))) ( -31.20) >DroSim_CAF1 80717 115 - 1 CUUGGAAACAAGGGCUUGCAGUUGGCGUGUUGCACGACUUAAGCUUGAAGCCGAGUUUUA----AA-AAAGCAGUAUACUUAAUUUUUAUUUAGCAACAUGUAAGUUAUAACAUAACUGU ((((....)))).....((((((((((((((((.......(((((((....)))))))((----((-((...((....))...))))))....)))))))))..((....)).))))))) ( -30.10) >DroEre_CAF1 86336 118 - 1 CUUGGAAACAAGGGGUUGCAGUUGGCGUGUUGCACGACUUAAAUUUAAAGGCGAAUUUAAAAUGAA-AAAACAGAGUACUUAAUUUUUAAUUUGCAACAUGUAAGUUAUAACAUAACUG- ((((....))))......((((((((((((((((....((((((((......))))))))..((((-((...((....))...))))))...))))))))))..((....)).))))))- ( -26.20) >DroYak_CAF1 82947 115 - 1 CUUGGAAACAAUGGCUUGCAGUUGGCGUGUUGCACGACUUAAAUUUAAAGGCGAAUUUUA----AA-AAAGCAAAGUACUUAAUUUUUAAUUUGCAACAUAUAAGUUACAACAUAACUGU .(((....)))......((((((...((((((...((((((..(((((((.....)))))----))-...(((((...............)))))......)))))).)))))))))))) ( -22.16) >consensus CUUGGAAACAAGGGCUUGCAGUUGGCGUGUUGCACGACUUAAACUUGAAGCCGAGUUUUA____AA_AAAGCAGUAUACUUAAUUUUUAAUUAGCAACAUGUAAGUUAUAACAUAACUGU ((((....)))).....((((((((((((((((.......(((((((....)))))))..............((....)).............)))))))))..((....)).))))))) (-21.16 = -22.12 + 0.96)



| Location | 27,766,060 – 27,766,160 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27766060 100 - 27905053 AGCCUCAUGGUUGUUUUAGUUUAUACCUCGGAAACAAGGGCUUGCAGUUGGCGUGUUGCACGACUUAAACUUGUAGCCGAGUCUAA----AAAAAAACAGU--ACU ((((....))))(((((..((((...(((((..(((((...(((.(((((((.....)).)))))))).)))))..)))))..)))----)..)))))...--... ( -27.50) >DroSec_CAF1 80455 100 - 1 AGC-UGUUGGUUGUUUCAGUUUAUACCUUGGAAACAAGAGCUUGCAGUUGGCGUGUUGCACGACUUAAACUUGAAGCCGAGUUUUA----AA-AAAGCAGUAUACU .((-((((((((((....((..((((((((....)))).(((.......))))))).)))))))).(((((((....)))))))..----..-..))))))..... ( -25.10) >DroSim_CAF1 80757 100 - 1 AGC-UGUUGGUUGUUUCGGUUUAUACCUUGGAAACAAGGGCUUGCAGUUGGCGUGUUGCACGACUUAAGCUUGAAGCCGAGUUUUA----AA-AAAGCAGUAUACU .((-((((..(((.((((((((...(((((....)))))(((((.(((((((.....)).))))))))))...))))))))...))----).-..))))))..... ( -38.20) >DroEre_CAF1 86375 104 - 1 AGC-UCUUGGUUGUUUCGGCUUAUACCUUGGAAACAAGGGGUUGCAGUUGGCGUGUUGCACGACUUAAAUUUAAAGGCGAAUUUAAAAUGAA-AAAACAGAGUACU .((-(((..(((.(((((((.....(((((....)))))....)).((((((.....)).))))((((((((......))))))))..))))-).))))))))... ( -28.40) >DroYak_CAF1 82987 100 - 1 AGC-UCUUCGUUGUUUCGGUUUAUACCUUGGAAACAAUGGCUUGCAGUUGGCGUGUUGCACGACUUAAAUUUAAAGGCGAAUUUUA----AA-AAAGCAAAGUACU ...-....((((((((((((....)))...)))))))))(((((((((((((.....)).)))))....(((((((.....)))))----))-...)).))))... ( -22.90) >consensus AGC_UCUUGGUUGUUUCGGUUUAUACCUUGGAAACAAGGGCUUGCAGUUGGCGUGUUGCACGACUUAAACUUGAAGCCGAGUUUUA____AA_AAAGCAGUAUACU ..........((((((..((.....(((((....)))))....))(((((((.....)).))))).(((((((....)))))))..........))))))...... (-18.02 = -18.02 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:16 2006