
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,728,566 – 27,728,674 |
| Length | 108 |
| Max. P | 0.686255 |

| Location | 27,728,566 – 27,728,661 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.05 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27728566 95 - 27905053 UAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGUGGUCGCUUCAAUAUAAAUGCAGUUACAAAUGGUAGACG--C---CUCCU-------------------- .........((((..(((((....))))).........(((((.....)))))))))............((..((((.....))))..)--)---.....-------------------- ( -18.40) >DroGri_CAF1 49465 94 - 1 UAUCUGCGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAAUGAUG-CCU--C---AUCCC-------------------- ...((((..((((..(((((....))))).........(((((.....)))))))))....((....))))))........((((-...--)---)))..-------------------- ( -19.90) >DroWil_CAF1 48152 94 - 1 UAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAAUGGCUGCCA--U---AUGC--------------------- ....((((.((((..(((((....))))).........(((((.....)))))))))............(((((((....)))))))))--)---)...--------------------- ( -25.00) >DroMoj_CAF1 49998 94 - 1 UAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAAUGAUG-CCG--A---CAGAC-------------------- ..(((((.(((((..(((((....))))).........(((((.....)))))))))............(((...........))-).)--)---)))).-------------------- ( -23.20) >DroAna_CAF1 43089 94 - 1 UAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUUAAUAUAAAUGCAGUUGCGAAUGUCG-GCC--C---GCCCU-------------------- ........(((((...(((((((..............((((((.....))))))...........))))))).)))))......(-(..--.---.))..-------------------- ( -22.01) >DroPer_CAF1 48321 120 - 1 UAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAAUGGCACAGUUUCGUACUCCCUCUCCGCAUACUGAGAUCGG ...((((((((((..(((((....))))).........(((((.....))))))))).............((........)))))))).........((.((((........))))..)) ( -28.90) >consensus UAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAAUGGCG_CCG__C___CUCCC____________________ ...((((..((((..(((((....))))).........(((((.....)))))))))....((....))))))............................................... (-17.33 = -17.05 + -0.28)

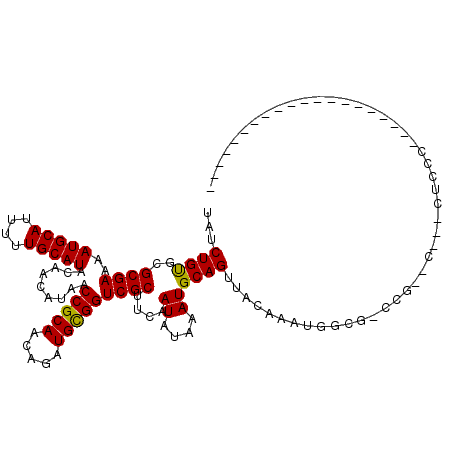

| Location | 27,728,581 – 27,728,674 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.17 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27728581 93 - 27905053 ---------------------------UGUUUUGCAUGUUUAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGUGGUCGCUUCAAUAUAAAUGCAGUUACAAA ---------------------------(((.((((((...(((.((...((((..(((((....))))).........(((((.....)))))))))..)))))...))))))..))).. ( -23.50) >DroVir_CAF1 49083 93 - 1 ---------------------------AGUUCUGUCUGCUUAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAA ---------------------------.((.((((..((........))((((..(((((....))))).........(((((.....)))))))))............))))..))... ( -22.80) >DroGri_CAF1 49479 93 - 1 ---------------------------AGUUCUGUUUGUUUAUCUGCGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAA ---------------------------.((.((((..((((((......((((..(((((....))))).........(((((.....)))))))))......))))))))))..))... ( -23.00) >DroMoj_CAF1 50012 93 - 1 ---------------------------CGUUCUGUUUGCUUAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAA ---------------------------.((.((((..((........))((((..(((((....))))).........(((((.....)))))))))............))))..))... ( -22.90) >DroAna_CAF1 43103 93 - 1 ---------------------------AGUUUUGUAUGUUUAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUUAAUAUAAAUGCAGUUGCGAA ---------------------------....((((((((......(((((......)))))....)))))))).....(((((.....)))))((((..................)))). ( -22.47) >DroPer_CAF1 48361 120 - 1 GCGGGGAGUCGAGGGGCGUCGCGGGAGUUUUCUGUGUGUUUAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAA (((.(((......(((((.(((((((...)))))))))))).))).)))((((..(((((....))))).........(((((.....)))))))))....................... ( -32.40) >consensus ___________________________AGUUCUGUAUGUUUAUCUGUGCGCGAAAAUGCAUUUUUGCAUACAACAUAACCGCAACAGAUGCGGUCGCUUCAAUAUAAAUGCAGUUACAAA ............................((.((((..((........))((((..(((((....))))).........(((((.....)))))))))............))))..))... (-20.86 = -20.17 + -0.69)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:07 2006