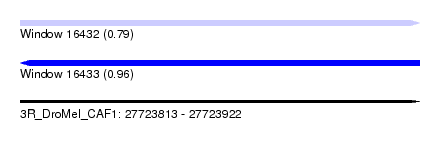
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,723,813 – 27,723,922 |
| Length | 109 |
| Max. P | 0.959097 |

| Location | 27,723,813 – 27,723,922 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27723813 109 + 27905053 GG-AAA-AAAACAUGUUUAAUGCCCCGCCAUUUUUUAUAGCGCCACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAU-UUAGCACAUUAUUUGC--CGGGC------ ..-...-..............((((.((..........(((((...........)))))((((((..((.((........)).))..)))-)))...........))--.))))------ ( -16.40) >DroVir_CAF1 43937 98 + 1 ---------AAAAUUUUUAAUGCCACGCCAUUUUUUAUGGCGCCACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAU-UUAGCGCAUUAUUUUU--U---------- ---------................((((((.....)))))).........((((((((.(((((..((.((........)).))..)))-))))))))))......--.---------- ( -22.00) >DroGri_CAF1 44410 98 + 1 ---------AAAAUUUUUAAUGCCACGCCAUUUUUUAUGGCGCCACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAU-UUAGCGCAUUAUAUUU--U---------- ---------................((((((.....)))))).........((((((((.(((((..((.((........)).))..)))-))))))))))......--.---------- ( -22.00) >DroMoj_CAF1 44305 108 + 1 ---------AAAAUUUUUAAUGCCACGCCAUUUUUUAUGGCGCCACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAU-UUAGCGCAUUAUUUUC--CAGGCAGCGAC ---------...........((((.((((((.....)))))).........((((((((.(((((..((.((........)).))..)))-))))))))))......--..))))..... ( -28.70) >DroAna_CAF1 38076 109 + 1 GA-AAGGAAAACAU-UUUAAUGCCCCGCCAUUU-UUAUGGCGCCACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAUUUUAGCGCAUUAUUCGC--CGGGC------ ..-...........-......((((((((((..-..)))))).........((((((((.(((((..((.((........)).))..))))).))))))))......--.))))------ ( -29.00) >DroPer_CAF1 42639 111 + 1 AACAAA--AAAAUCUCUUAAUGCCCCGCCAUUUUUUAUAGCGCUACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAU-UUAGCGCAUUAUUUGCGCAGGAC------ ......--......((((((((......)))).......((((........((((((((.(((((..((.((........)).))..)))-))))))))))....)))))))).------ ( -20.80) >consensus _________AAAAUUUUUAAUGCCACGCCAUUUUUUAUGGCGCCACACUUGAAUGCGCUUAAAUAUUUUAUGUAAUUUUUCGCAAUUUAU_UUAGCGCAUUAUUUGC__CGGGC______ .........................((((((.....)))))).........((((((((...(((..((.((........)).))..)))...))))))))................... (-17.22 = -17.72 + 0.50)


| Location | 27,723,813 – 27,723,922 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -21.93 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27723813 109 - 27905053 ------GCCCG--GCAAAUAAUGUGCUAA-AUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUGGCGCUAUAAAAAAUGGCGGGGCAUUAAACAUGUUUU-UUU-CC ------((((.--(((...((((((((((-(((...((..(....)..)).....))))).))))))))....)))..((((((.....))))))))))..............-...-.. ( -26.30) >DroVir_CAF1 43937 98 - 1 ----------A--AAAAAUAAUGCGCUAA-AUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUGGCGCCAUAAAAAAUGGCGUGGCAUUAAAAAUUUU--------- ----------.--......((((((((((-(((...((..(....)..)).....))))).))))))))..(((((.(((((((.....))))))).))))).........--------- ( -27.80) >DroGri_CAF1 44410 98 - 1 ----------A--AAAUAUAAUGCGCUAA-AUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUGGCGCCAUAAAAAAUGGCGUGGCAUUAAAAAUUUU--------- ----------.--......((((((((((-(((...((..(....)..)).....))))).))))))))..(((((.(((((((.....))))))).))))).........--------- ( -27.80) >DroMoj_CAF1 44305 108 - 1 GUCGCUGCCUG--GAAAAUAAUGCGCUAA-AUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUGGCGCCAUAAAAAAUGGCGUGGCAUUAAAAAUUUU--------- (((((.(((..--......((((((((((-(((...((..(....)..)).....))))).)))))))).......)))(((((.....))))))))))............--------- ( -29.06) >DroAna_CAF1 38076 109 - 1 ------GCCCG--GCGAAUAAUGCGCUAAAAUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUGGCGCCAUAA-AAAUGGCGGGGCAUUAAA-AUGUUUUCCUU-UC ------((((.--(((...((((((((.(((((...((..(....)..)).....))))).))))))))....)))..((((((..-..))))))))))......-...........-.. ( -30.60) >DroPer_CAF1 42639 111 - 1 ------GUCCUGCGCAAAUAAUGCGCUAA-AUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUAGCGCUAUAAAAAAUGGCGGGGCAUUAAGAGAUUUU--UUUGUU ------(((((((((....((((((((((-(((...((..(....)..)).....))))).))))))))...))))).((((((.....)))))))))).............--...... ( -27.20) >consensus ______GCCCG__GAAAAUAAUGCGCUAA_AUAAAUUGCGAAAAAUUACAUAAAAUAUUUAAGCGCAUUCAAGUGUGGCGCCAUAAAAAAUGGCGGGGCAUUAAAAAUUUU_________ ...................((((((((...(((...((..(....)..)).....)))...))))))))..(((((..((((((.....))))))..))))).................. (-21.93 = -21.57 + -0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:04 2006