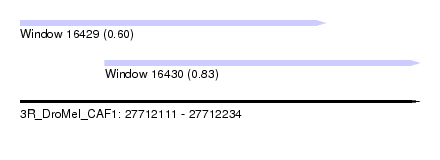
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,712,111 – 27,712,234 |
| Length | 123 |
| Max. P | 0.831731 |

| Location | 27,712,111 – 27,712,205 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27712111 94 + 27905053 -------CCAGGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCAGCCACAAAAGUCGGAGGAGGAGUACCGAAA -------...((((................))))(((..(((......)))..)))(((((((...((((..((((....)).)).)))).)))).))).. ( -21.89) >DroPse_CAF1 39161 86 + 1 GAAGGCUGAAGGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUGUAACAGCUGCGGUUCCUGCCACAAUCA---------------AGGAGCCCCAGAGAAA ...(((....((((................)))))))............(((.(((((((..........---------------))))))).)))..... ( -21.19) >DroSec_CAF1 34920 94 + 1 -------CUAGGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGGAGGAGUACCGAAA -------...((((................))))(((..(((......)))..)))(((((((...(((((((.......)))))))....)))).))).. ( -25.79) >DroSim_CAF1 35065 94 + 1 -------CUAGGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGGAGGAGUACCGAAA -------...((((................))))(((..(((......)))..)))(((((((...(((((((.......)))))))....)))).))).. ( -25.79) >DroEre_CAF1 35584 94 + 1 -------CUGGGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUAUAACAGCUGCGGUUCGUGCUAAACUCCGGCCGCAAAAGUCGGAGGAGGAGUAUCGAAA -------...((((................))))(((..(((......)))..)))(((((((...(((((((.......)))))))....)))).))).. ( -25.29) >DroYak_CAF1 36426 94 + 1 -------CUAAGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGGAGGAGUAUCGAAA -------........................(((((...(((......))))))))(((((((...(((((((.......)))))))....)))).))).. ( -25.10) >consensus _______CUAGGCAAUAAAUUAUAUGAAAAUGCCGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGGAGGAGUACCGAAA ...............................(((((...(((......))))))))(((((((...(((((((.......)))))))....))))).)).. (-19.91 = -20.45 + 0.54)
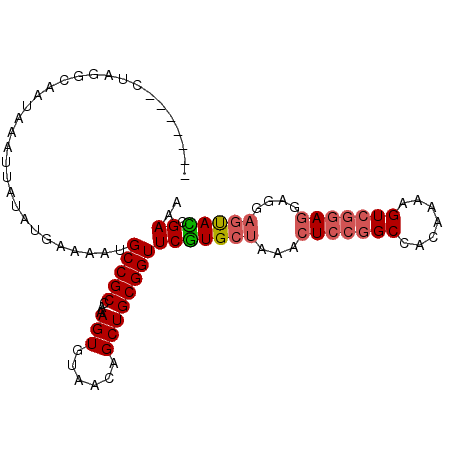

| Location | 27,712,137 – 27,712,234 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -24.51 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27712137 97 + 27905053 CGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCAGCCACAAAAGUCGGAGG--AGGAGUACCGAAACCACAGCAACAAGAAAGAGUCACUACUGG ..(((.((((.....((((.((((((((((...((((..((((....)).)).))--)).)))).)).)))).)))).((........))))))..))) ( -27.90) >DroSec_CAF1 34946 94 + 1 CGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGG--AGGAGUACCGAAACCACAGCAACAAGAAAGAGUCAC---UGA ......((((.....((((.((((((((((...(((((((.......))))))).--...)))).)).)))).)))).((........)))))---).. ( -30.40) >DroSim_CAF1 35091 94 + 1 CGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGG--AGGAGUACCGAAACCACAGCAACAAGAAAGAGUCAC---UGA ......((((.....((((.((((((((((...(((((((.......))))))).--...)))).)).)))).)))).((........)))))---).. ( -30.40) >DroEre_CAF1 35610 94 + 1 CGCCAAAGUAUAACAGCUGCGGUUCGUGCUAAACUCCGGCCGCAAAAGUCGGAGG--AGGAGUAUCGAAACCACAGCAACAAGAAAGAGUCAC---UGG ..(((..........((((.((((((((((...(((((((.......))))))).--...)))).)).)))).)))).....((.....))..---))) ( -29.80) >DroYak_CAF1 36452 94 + 1 CGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGG--AGGAGUAUCGAAACCACAGCAACAAGAAAGAGUCAC---UGG ......((((.....((((.((((((((((...(((((((.......))))))).--...)))).)).)))).)))).((........)))))---).. ( -30.40) >DroAna_CAF1 35802 89 + 1 CGCCAAAGCGUAGCAGCUGCGGUUCUUGCCGAAC-------ACAGGAGCUGAGGUACGGGGGCACGGAAACCACAGCAACAAGAAAGAGUCGC---CGG ..((..(((......)))(((..((((.((....-------...)).((((.(((.((......))...))).)))).......))))..)))---.)) ( -26.30) >consensus CGCCAAAGUGUAACAGCUGCGGUUCGUGCUAAACUCCGGCCACAAAAGUCGGAGG__AGGAGUACCGAAACCACAGCAACAAGAAAGAGUCAC___UGG ...............((((.((((((((((...(((((((.......)))))))......)))).)).)))).))))...................... (-24.51 = -24.98 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:54:02 2006