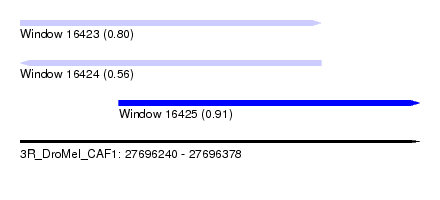
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,696,240 – 27,696,378 |
| Length | 138 |
| Max. P | 0.906117 |

| Location | 27,696,240 – 27,696,344 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.62 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27696240 104 + 27905053 CACCAAACACAUGCAAACACAUGCAAACAACAAGCGCUUAGA-AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGGGGAUCUUGA---CU-AAAUGGAGACAUCCAUAU ..((.......((((......)))).......((.(((((..-..((((....)))).))))).))........))((((.((..---(.-....)..)).)))).... ( -19.40) >DroSec_CAF1 18733 100 + 1 CGCCCGA---GUGCAAACACAUGCAAACAACAAGCGCUUGGA-AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGGAUCUUGAAAACUGAAAGGGAGA--UUAA--- ..(((.(---((.(((...(((((((((.....).(((((..-...))))).))))))))..((((.......))))....)))...)))....)))...--....--- ( -23.30) >DroSim_CAF1 18837 100 + 1 CGCCCGA---GUGCAAACACAUGCAAACAACAAGCGCUUGGA-AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGGAUCUUGAAAACUGAAAGGGAGA--UUAA--- ..(((.(---((.(((...(((((((((.....).(((((..-...))))).))))))))..((((.......))))....)))...)))....)))...--....--- ( -23.30) >DroEre_CAF1 18956 88 + 1 ----------------ACACACACAAACAACCAGCGCUUGAAAAAGCGAGCGUUUGCAUGAGCACUGAGAGAAAGUGCAUCUCGAAAAGUAAAAGGGACU--CUAA--- ----------------...((..(((((......(((((....)))))...)))))..)).(((((.......))))).((((...........))))..--....--- ( -20.60) >DroYak_CAF1 19243 101 + 1 CAGCCCA---GUGCACACACAUGCAAACAACAAGCGCUUGGAAAAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGCAUCUCGAAACCCAAAUGGGAGA--CUAU--- ...((((---.........(((((((((.....).(((((......))))).)))))))).(((((.......)))))...............))))...--....--- ( -26.40) >consensus CACCCGA___GUGCAAACACAUGCAAACAACAAGCGCUUGGA_AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGGAUCUUGAAAACUAAAAGGGAGA__UUAA___ ..(((..............(((((((((.....).(((((......))))).))))))))..((((.......)))).................)))............ (-13.42 = -14.62 + 1.20)



| Location | 27,696,240 – 27,696,344 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -15.38 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27696240 104 - 27905053 AUAUGGAUGUCUCCAUUU-AG---UCAAGAUCCCCUUUUUCUCAGUGCUCAUGCAAAUGCUUGCUU-UCUAAGCGCUUGUUGUUUGCAUGUGUUUGCAUGUGUUUGGUG ..(((((....)))))..-..---.................((((.((.(((((((((((.(((..-....(((....)))....))).))))))))))).)))))).. ( -24.10) >DroSec_CAF1 18733 100 - 1 ---UUAA--UCUCCCUUUCAGUUUUCAAGAUCCACUUUUUCUCAGUGCUCAUGCAAAUGCUUGCUU-UCCAAGCGCUUGUUGUUUGCAUGUGUUUGCAC---UCGGGCG ---....--...(((............(((.........))).(((((.((((((((((((((...-..))))).......))))))))).....))))---).))).. ( -22.71) >DroSim_CAF1 18837 100 - 1 ---UUAA--UCUCCCUUUCAGUUUUCAAGAUCCACUUUUUCUCAGUGCUCAUGCAAAUGCUUGCUU-UCCAAGCGCUUGUUGUUUGCAUGUGUUUGCAC---UCGGGCG ---....--...(((............(((.........))).(((((.((((((((((((((...-..))))).......))))))))).....))))---).))).. ( -22.71) >DroEre_CAF1 18956 88 - 1 ---UUAG--AGUCCCUUUUACUUUUCGAGAUGCACUUUCUCUCAGUGCUCAUGCAAACGCUCGCUUUUUCAAGCGCUGGUUGUUUGUGUGUGU---------------- ---..((--(((.......)))))..((((.......)))).....((.(((((((((((((((((....)))))..)).)))))))))).))---------------- ( -20.20) >DroYak_CAF1 19243 101 - 1 ---AUAG--UCUCCCAUUUGGGUUUCGAGAUGCACUUUUUCUCAGUGCUCAUGCAAAUGCUUGCUUUUCCAAGCGCUUGUUGUUUGCAUGUGUGUGCAC---UGGGCUG ---.(((--((((((....)))....((((.........))))(((((.((((((...(((((......)))))((.........)).)))))).))))---))))))) ( -31.80) >consensus ___UUAA__UCUCCCUUUCAGUUUUCAAGAUCCACUUUUUCUCAGUGCUCAUGCAAAUGCUUGCUU_UCCAAGCGCUUGUUGUUUGCAUGUGUUUGCAC___UCGGGCG ..............................................((.(((((((((((.(((((....)))))...).)))))))))).))................ (-15.38 = -14.74 + -0.64)


| Location | 27,696,274 – 27,696,378 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.55 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -8.92 |
| Energy contribution | -8.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27696274 104 + 27905053 CGCUUAGA-AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGGGGAUCUUGA---CU-AAAUGGAGACAUCCAUAUAUGUAAAUGUCUAAAUACGGCAAAGAUUUUAUCA------ .(((((..-..((((....)))).)))))............((((((((.(---((-(.(((((....))))).)).))...((((.......))))))))))))....------ ( -20.10) >DroSec_CAF1 18764 98 + 1 CGCUUGGA-AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGGAUCUUGAAAACUGAAAGGGAGA--UUAA----------UG----CAUCCAGCCCUAGUUUUAUAAGAAGCU .(((((..-...)))))....((.....((((.......))))..(((((((((((...((((.((--(...----------..----.)))...)))))))))).))))).)). ( -24.20) >DroSim_CAF1 18868 98 + 1 CGCUUGGA-AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGGAUCUUGAAAACUGAAAGGGAGA--UUAA----------UG----CAUCCAGCCCUAGUUUUAUAAGAAGCU .(((((..-...)))))....((.....((((.......))))..(((((((((((...((((.((--(...----------..----.)))...)))))))))).))))).)). ( -24.20) >DroEre_CAF1 18974 99 + 1 CGCUUGAAAAAGCGAGCGUUUGCAUGAGCACUGAGAGAAAGUGCAUCUCGAAAAGUAAAAGGGACU--CUAA----------UG----CAUAGGGUCCCACUUUUAUUUGAAGCC (((((....))))).((.((((...(((((((.......))))).)).))))((((((((((((((--(((.----------..----..)))))))))..))))))))...)). ( -34.00) >DroYak_CAF1 19274 99 + 1 CGCUUGGAAAAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGCAUCUCGAAACCCAAAUGGGAGA--CUAU----------UG----UGUUCAGUCCUACUUUAAUUAGAAGCA .(((((((...((((....)))).((((((((((((.........))))).......(((((....--))))----------))----)))))).)))..((......)))))). ( -25.90) >consensus CGCUUGGA_AAGCAAGCAUUUGCAUGAGCACUGAGAAAAAGUGGAUCUUGAAAACUAAAAGGGAGA__UUAA__________UG____CAUACAGCCCUACUUUUAUAAGAAGCU .(((((......)))))....((.....((((.......))))..((((...........))))..............................))................... ( -8.92 = -8.96 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:53:57 2006