
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,677,687 – 27,677,936 |
| Length | 249 |
| Max. P | 0.939538 |

| Location | 27,677,687 – 27,677,786 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.21 |
| Mean single sequence MFE | -16.96 |
| Consensus MFE | -12.27 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
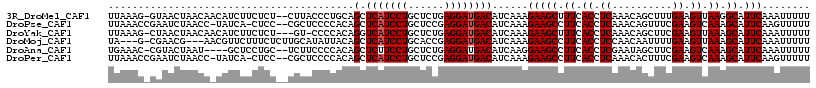
>3R_DroMel_CAF1 27677687 99 - 27905053 AAUUU--UU--UUUCCCUUCUAUGUGUGCUAUUAAAACU----AUCGGAGCAACCACUAAUAACCUCGUCCUUUUUAUUUUACAGGAAGGACCGUAAAAUGGCACUG .....--..--..............((((((((......----...((.....))............(((((((((.......))))))))).....)))))))).. ( -19.50) >DroSec_CAF1 160 102 - 1 -UUUUUUAUUUUGCCAUUUGUAUGUGUGCUAUUAAAACU----AUCGGACCUACCACUAAAAACCUCAUCCUUUUUAUUUUUCAGGAAGGACCGUAAAAUGGCACUG -..........((((((((.((((.((.((.........----...((.....)).............((((...........)))))).))))))))))))))... ( -18.60) >DroEre_CAF1 153 82 - 1 ---------------------AUAUGAGCUGUUAAAACUCUUUAACGGAAUCACCACUAA-AACCGUAUCCUUAU---CACACAGGAAAGACCGAAAAAUGGCACUG ---------------------......((((((.....((((..((((............-..)))).((((...---.....))))))))......)))))).... ( -11.14) >DroYak_CAF1 153 83 - 1 ---------------------AUGUGUGCUAUUAAAACUGCUUAACGGACUUACCACUAAAAACCGCAUCCUUUC---CCUACAGGAAGGACCGAAAAAUGGCACUG ---------------------....((((((((.............((.....)).........((..(((((((---......))))))).))...)))))))).. ( -18.60) >consensus _____________________AUGUGUGCUAUUAAAACU____AACGGACCUACCACUAAAAACCGCAUCCUUUU___CUUACAGGAAGGACCGAAAAAUGGCACUG .........................((((((((.............((.....)).............((((((((.......))))))))......)))))))).. (-12.27 = -13.15 + 0.88)



| Location | 27,677,786 – 27,677,902 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27677786 116 + 27905053 UUAAAAAUAGUUGUCUGUGC-GGCCAUUACUUACGGGAAAAAUUUGAAUGCCUUAACUUCAAAGCUGUUUGAGGUGAAAGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGAGCUGCA ......((((....))))((-(((((((..............((((((.(......))))))).(((((((((((((...(......)...))).))))))).))))))).))))). ( -29.60) >DroVir_CAF1 223 112 + 1 -----AAUUGUUAUCUUUAUAUUCAUUUACUUACGGGAAAAAUUUGAAUGCUUUAACCUCGAAAUUGUUGGAGCUGAAGGCUUCUUUGAUGUCAUCCUCGGUACAGGAUGAGCUGUA -----..........................(((((.........(((.(((((...(((.((....)).)))...)))))))).......(((((((......))))))).))))) ( -21.50) >DroSec_CAF1 262 115 + 1 -AAAAAUUAGUUGUCUGUGC-GGGUUUUACUUACGGGAAAAAUUUGAAUGCUUUAACUUCGAAGCUGUUUGAGGUGAAAGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGAGCUGCA -................(((-((......................(((.(((((.((((((((....)))))))).)))))))).......(((((((......))))))).))))) ( -28.10) >DroWil_CAF1 230 108 + 1 ----AGAAGGCUGAUUCUG-----CAUUACUUACGGGAAAAAUUUGAAUGCUUUGACUUCGAAACUGUUCGAGGUGAAGGCCUCUUUGAUGUCAUCCUCAGAGCAGGAUGAGCUGUA ----....((((.((((((-----(....((...((((.....(..((.(((((.((((((((....)))))))).)))))...))..).....)))).)).))))))).))))... ( -34.80) >DroYak_CAF1 236 98 + 1 ------------------GC-UGCCUUUACUUACGGGAAAAAUUUGAAUGCUUUAACUUCGAAGCUGUUUGAGGUGAAAGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGACCUGUG ------------------..-..........((((((........(((.(((((.((((((((....)))))))).)))))))).......(((((((......))))))))))))) ( -28.90) >DroMoj_CAF1 215 112 + 1 -----AAUUUUGAAAAUCAUUAAUAUUAACUUACGGGAAAAAUUUGAAUGCUUUAACUUCAAAAUUGUUGGAGGUGAAGGCUUCUUUGAUGUCAUCCUCGGUGCAGGAUGAGCUGUA -----..........................(((((.........(((.(((((..((((((.....))))))...)))))))).......(((((((......))))))).))))) ( -23.80) >consensus _____AAUAGUUGUCUGUGC_GGCCUUUACUUACGGGAAAAAUUUGAAUGCUUUAACUUCGAAACUGUUUGAGGUGAAAGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGAGCUGUA ...............................(((((.........(((.(((((.((((((((....)))))))).)))))))).......(((((((......))))))).))))) (-22.58 = -23.13 + 0.56)

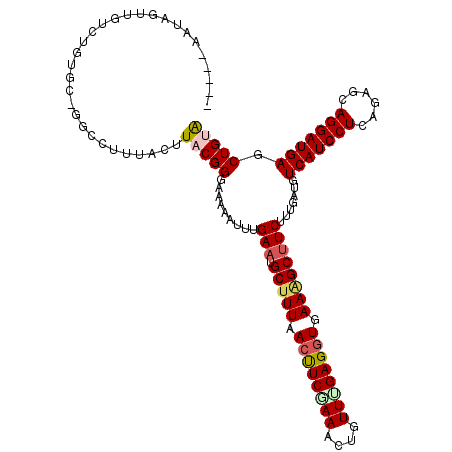

| Location | 27,677,822 – 27,677,936 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
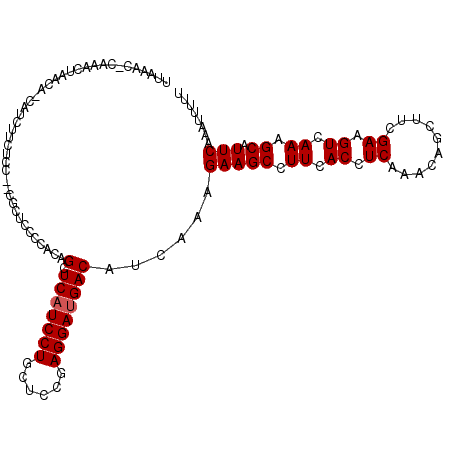
>3R_DroMel_CAF1 27677822 114 + 27905053 AAAAAUUUGAAUGCCUUAACUUCAAAGCUGUUUGAGGUGAAAGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGAGCUGCAGGGUAAG--AGAGAAGAUGUUGUUAGUUAC-CUUUAA .....((((((.(......))))))).......(((((((.(((.(((((..(.(.((((((((..(.....)..))).))))).).--)..))))).))).....))))-)))... ( -25.10) >DroPse_CAF1 262 113 + 1 AAAAACUUGAAUGCUUUGACUUCGAAACUGUUUGAGGUGAAGGCUUCUUUGAUGUCAUCCUCGGAGCAGGAUGAGCUGUGGGGAGCG--GGAG-UGAUA-GGUUAGAUUCGGUUUAA ..(((((.(((.(((((.((((((((....)))))))).))))).(((....((((.((((((.(((.......))).))))))((.--...)-)))))-....))))))))))).. ( -36.20) >DroYak_CAF1 254 112 + 1 AAAAAUUUGAAUGCUUUAACUUCGAAGCUGUUUGAGGUGAAAGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGACCUGUGGGG-AC---AGAGAAGAUGUUGUUAGUUAG-CUUUAA ........(((.(((((.((((((((....)))))))).))))))))......((((((((......))))))))((.((...-.)---).)).....((((.....)))-)..... ( -31.60) >DroMoj_CAF1 247 110 + 1 AAAAAUUUGAAUGCUUUAACUUCAAAAUUGUUGGAGGUGAAGGCUUCUUUGAUGUCAUCCUCGGUGCAGGAUGAGCUGUAAUAUGCAAGAGAAAGAACGUU---CGUUCG-C---UA ............(((((..((((((.....))))))...)))))((((((....(((((((......)))))))((........)).)))))).((((...---.)))).-.---.. ( -27.70) >DroAna_CAF1 238 110 + 1 AAAAAUUUGAAUGCUUUGACUUCGAAGCUAUUCGAGGUGAAGGCUUCCUUGAUGUCAUCCUCAGAGCAGGAAGAGCUGUGGGGAAGA--GCAGGAGC----AUUAGUACG-GUUUCA .......((((.(((((.((((((((....)))))))).)))))(((((.(...((.((((((.(((.......))).)))))).))--.)))))).----.........-..)))) ( -36.60) >DroPer_CAF1 261 113 + 1 AAAAACUUGAAUGCUUUGACUUCGAAAGUGUUUGAGGUGAAGGCUUCUUUGAUGUCAUCCUCGGAGCAGGAUGAGCUGUGGGGAGCG--GGAG-UGAUA-GGUUAGAUUCGGUUUAA ..(((((.(((.(((((.((((((((....)))))))).))))).(((....((((.((((((.(((.......))).))))))((.--...)-)))))-....))))))))))).. ( -36.20) >consensus AAAAAUUUGAAUGCUUUAACUUCGAAACUGUUUGAGGUGAAGGCUUCUUUGAUGUCAUCCUCAGAGCAGGAUGAGCUGUGGGGAACG__AGAGAAGAUG_UGUUAGUUAC_CUUUAA ........(((.(((((.((((((((....)))))))).)))))))).......(((((((......)))))))........................................... (-22.18 = -22.35 + 0.17)
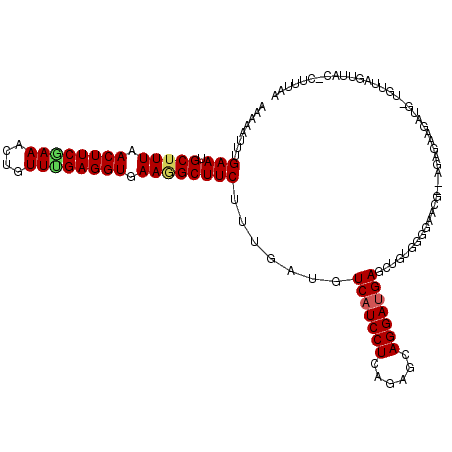
| Location | 27,677,822 – 27,677,936 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -20.00 |
| Consensus MFE | -11.78 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27677822 114 - 27905053 UUAAAG-GUAACUAACAACAUCUUCUCU--CUUACCCUGCAGCUCAUCCUGCUCUGAGGAUGACAUCAAAGAAGCUUUCACCUCAAACAGCUUUGAAGUUAAGGCAUUCAAAUUUUU .....(-((((.................--.))))).....(.(((((((......))))))))......((((((((.((.(((((....))))).)).))))).)))........ ( -21.57) >DroPse_CAF1 262 113 - 1 UUAAACCGAAUCUAACC-UAUCA-CUCC--CGCUCCCCACAGCUCAUCCUGCUCCGAGGAUGACAUCAAAGAAGCCUUCACCUCAAACAGUUUCGAAGUCAAAGCAUUCAAGUUUUU .......((((((....-.....-....--.(((......)))(((((((......))))))).......(((((..............)))))........)).))))........ ( -15.54) >DroYak_CAF1 254 112 - 1 UUAAAG-CUAACUAACAACAUCUUCUCU---GU-CCCCACAGGUCAUCCUGCUCUGAGGAUGACAUCAAAGAAGCUUUCACCUCAAACAGCUUCGAAGUUAAAGCAUUCAAAUUUUU .....(-((...((((..........((---((-....))))((((((((......))))))))......((((((............))))))...)))).)))............ ( -25.70) >DroMoj_CAF1 247 110 - 1 UA---G-CGAACG---AACGUUCUUUCUCUUGCAUAUUACAGCUCAUCCUGCACCGAGGAUGACAUCAAAGAAGCCUUCACCUCCAACAAUUUUGAAGUUAAAGCAUUCAAAUUUUU ..---(-(((((.---...))))....((((((........))(((((((......))))))).....)))).))................((((((((....)).))))))..... ( -19.40) >DroAna_CAF1 238 110 - 1 UGAAAC-CGUACUAAU----GCUCCUGC--UCUUCCCCACAGCUCUUCCUGCUCUGAGGAUGACAUCAAGGAAGCCUUCACCUCGAAUAGCUUCGAAGUCAAAGCAUUCAAAUUUUU ......-......(((----(((.....--...(((.((.(((.......))).)).)))((((.((...(((((.(((.....)))..))))))).)))).))))))......... ( -22.50) >DroPer_CAF1 261 113 - 1 UUAAACCGAAUCUAACC-UAUCA-CUCC--CGCUCCCCACAGCUCAUCCUGCUCCGAGGAUGACAUCAAAGAAGCCUUCACCUCAAACACUUUCGAAGUCAAAGCAUUCAAGUUUUU .......((((((....-..((.-....--.(((......)))(((((((......))))))).......)).(.((((...............)))).)..)).))))........ ( -15.26) >consensus UUAAAC_CAAACUAACA_CAUCUUCUCC__CGCUCCCCACAGCUCAUCCUGCUCCGAGGAUGACAUCAAAGAAGCCUUCACCUCAAACAGCUUCGAAGUCAAAGCAUUCAAAUUUUU .........................................(.(((((((......))))))))......(((((.((.((.((..........)).)).)).)).)))........ (-11.78 = -11.95 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:53:54 2006