
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,649,266 – 27,649,361 |
| Length | 95 |
| Max. P | 0.639130 |

| Location | 27,649,266 – 27,649,361 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.95 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
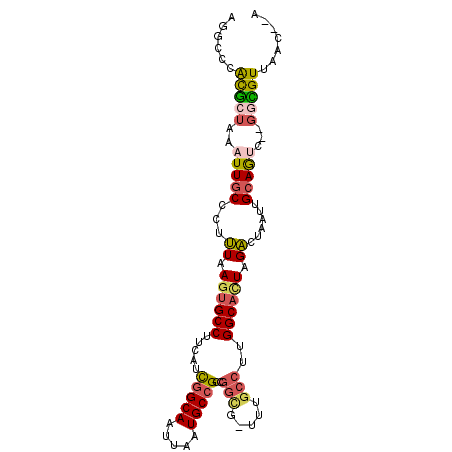
>3R_DroMel_CAF1 27649266 95 + 27905053 U--GUUAACGCC--GACUGCAAUUAGUCUAGUGCCAAGGCAAA-CGCCGGCGGCAUUAAUUGCCGAUGAAGGCACUUAAAGGGCAAUUUACCGUGGGCCU .--......(((--.((...((((.((((((((((..(((...-.)))..(((((.....))))).....))))))....))))))))....)).))).. ( -33.20) >DroPse_CAF1 36761 100 + 1 UUGGGUAACACACCGGUUGCAAUUAGUCUAAGGCCAAGGCAAAAAACUAGCGGCAUUAAUUGCCAAUGAAGGCACUUAAAAGGUAAUUUAGCAUCCGAGG ((((((.....(((.(((((...((((.....((....)).....))))))))).((((.((((......)))).))))..)))........)))))).. ( -23.22) >DroSec_CAF1 29456 95 + 1 U--GUUAACGCC--GACUGCAAUUAGUCUAGUGCCAAGGCAAA-CGCCGGCGGCAUUAAUUGCCGAUGAAGGCACUUAAAGGGCAAUUUAGCGUGGGCCU .--......(((--.((.((((((.((((((((((..(((...-.)))..(((((.....))))).....))))))....))))))))..)))).))).. ( -35.30) >DroSim_CAF1 34281 95 + 1 U--GUUAACGCC--GACUGCAAUUAGUCUAGUGCCAAGGCAAA-CGCCGGCGGCAUUAAUUGCCGAUGAAGGCACUUAAAGGGCAAUUUAGCGUGGGCCU .--......(((--.((.((((((.((((((((((..(((...-.)))..(((((.....))))).....))))))....))))))))..)))).))).. ( -35.30) >DroEre_CAF1 29576 95 + 1 U--GUUAACGCC--GACUGCAAUUAGUCUAGUGCCAAGGCAAA-AGCCGGCGGCAUUAAUUGCCAAUGAAGGCACUUAAAGGGCAAUUUAGCGUGGACCU .--(((((.(((--(((((....))))).((((((..(((...-.)))...((((.....))))......)))))).....)))...)))))........ ( -32.20) >DroAna_CAF1 28286 90 + 1 U--GUUAACAGC--GGCUGCAAUUAGCCAAGUGCCAAGGAGAG-UGGCGGCGGCAUUAAUUGCUGGUGAGGGCAGUUAGAG-GCAAUUUAGUGC----CG .--.......((--(((((....)))))...(((((.......-)))))))(((((((((((((..(((......)))..)-)))).)))))))----). ( -30.80) >consensus U__GUUAACGCC__GACUGCAAUUAGUCUAGUGCCAAGGCAAA_CGCCGGCGGCAUUAAUUGCCGAUGAAGGCACUUAAAGGGCAAUUUAGCGUGGGCCU .........(((..(((((....))))).((((((..(((.....)))..(((((.....))))).....)))))).....)))................ (-19.95 = -20.95 + 1.00)



| Location | 27,649,266 – 27,649,361 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27649266 95 - 27905053 AGGCCCACGGUAAAUUGCCCUUUAAGUGCCUUCAUCGGCAAUUAAUGCCGCCGGCG-UUUGCCUUGGCACUAGACUAAUUGCAGUC--GGCGUUAAC--A ..(((...(((.....))).....((((((.....(((((.....)))))..(((.-...)))..)))))).((((......))))--)))......--. ( -30.10) >DroPse_CAF1 36761 100 - 1 CCUCGGAUGCUAAAUUACCUUUUAAGUGCCUUCAUUGGCAAUUAAUGCCGCUAGUUUUUUGCCUUGGCCUUAGACUAAUUGCAACCGGUGUGUUACCCAA ..((((.(((..(((((...((((((.(((......(((((..(((.......)))..)))))..))))))))).)))))))).))))............ ( -24.20) >DroSec_CAF1 29456 95 - 1 AGGCCCACGCUAAAUUGCCCUUUAAGUGCCUUCAUCGGCAAUUAAUGCCGCCGGCG-UUUGCCUUGGCACUAGACUAAUUGCAGUC--GGCGUUAAC--A .(((....))).....(((.....((((((.....(((((.....)))))..(((.-...)))..)))))).((((......))))--)))......--. ( -30.00) >DroSim_CAF1 34281 95 - 1 AGGCCCACGCUAAAUUGCCCUUUAAGUGCCUUCAUCGGCAAUUAAUGCCGCCGGCG-UUUGCCUUGGCACUAGACUAAUUGCAGUC--GGCGUUAAC--A .(((....))).....(((.....((((((.....(((((.....)))))..(((.-...)))..)))))).((((......))))--)))......--. ( -30.00) >DroEre_CAF1 29576 95 - 1 AGGUCCACGCUAAAUUGCCCUUUAAGUGCCUUCAUUGGCAAUUAAUGCCGCCGGCU-UUUGCCUUGGCACUAGACUAAUUGCAGUC--GGCGUUAAC--A ..((..(((((.............((((((......((((.....))))...(((.-...)))..)))))).((((......))))--)))))..))--. ( -31.10) >DroAna_CAF1 28286 90 - 1 CG----GCACUAAAUUGC-CUCUAACUGCCCUCACCAGCAAUUAAUGCCGCCGCCA-CUCUCCUUGGCACUUGGCUAAUUGCAGCC--GCUGUUAAC--A .(----(((......)))-)..((((.((........(((((((..((((..((((-.......))))...)))))))))))....--)).))))..--. ( -22.80) >consensus AGGCCCACGCUAAAUUGCCCUUUAAGUGCCUUCAUCGGCAAUUAAUGCCGCCGGCG_UUUGCCUUGGCACUAGACUAAUUGCAGUC__GGCGUUAAC__A ......(((((..(((((...((.((((((.....(((((.....)))))..(((.....)))..)))))).))......)))))...)))))....... (-19.21 = -19.72 + 0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:53:48 2006