
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,626,772 – 27,626,886 |
| Length | 114 |
| Max. P | 0.523469 |

| Location | 27,626,772 – 27,626,886 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -16.87 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27626772 114 + 27905053 CCUCCAUUGUAAAACAGCUUGAGGGCGGCCUUGAACUGCAUCUAAGGACGUCCGAAGACGGAGGCAAUGACCUUGUUCAGUUUUUGAU---GCAAGAAAAGAGCUCCAUCAUAAUCA (((((.(((.....)))(((..(((((.((((((......)).)))).))))).)))..)))))..((((....((((..((((((..---.))))))..))))....))))..... ( -30.70) >DroPse_CAF1 7141 117 + 1 UGCCCGUUCUCAGCCAGUUGAAGGGCGGCGUGGAGCUGCACUUGCGAAGUUCGAAGGAUGGAGAUCACGAUUUUGUGCAGUUUCUCCUGAGGCAGGGCAAAUACUCGAUGAGUAUCA (((((..((((((((........)))((...((((((((((.........((....))..(((((....))))))))))))))).)))))))..))))).((((((...)))))).. ( -41.20) >DroSec_CAF1 6836 117 + 1 CCUCCAUUGUGGAGCAGCUUGAGGGCGGCCUUGAACUGCAUCUAAGGAGUUCGGAAGACGGAGGCAAUGAUCUUGUUCAGUUUUUGAUGGUGCAAGAAAAGAGCUCCAUCAAAAUUA (((((....((((((((.(..(((....)))..).)))).))))....((.(....))))))))...(((......)))..(((((((((.((.........)).)))))))))... ( -39.70) >DroSim_CAF1 6838 117 + 1 CCUCCAUUGUGGAGCAGUUUGAGGGCGGCCUUGAACUGCAUCUAAGGAGUUCGGAAGACGGAGGCAAUGAUCUUGUUCAGUUUUUGAUGGUGCAAGAAAAGAGCUCCAUCAAAAUUA (((((....((((((((((..(((....)))..)))))).))))....((.(....))))))))...(((......)))..(((((((((.((.........)).)))))))))... ( -43.90) >DroEre_CAF1 6794 117 + 1 GCUCCAUUGUACAACAGCUCGAGGGCGGCCUGGAACUGCAUCUAAGGAGCCCCGAGGACGGAGACCUCGAUCUUGUCCAGUUUUUGAUGGUGCAAGAAAAGAGCUCCAUAAAAAUAA .................((((.((((..((((((......))).))).))))))))...((((..(((..(((((((((........))).))))))...))))))).......... ( -37.70) >DroPer_CAF1 7157 117 + 1 UGCCCGUUCUCAGCCAGUUGAAGGGCGGCGUGGAGCUGCACUUGCGAAGUUCGAAGGAUGGAGAUCACGAUUUUGUGCGGUUUCUUCUGAGGCAGGGCAAAUACUCGAUGAGUAUCA (((((..((((((((.((......)))))..((((((((((.........((....))..(((((....)))))))))))))))...)))))..))))).((((((...)))))).. ( -39.40) >consensus CCUCCAUUGUAAAACAGCUUGAGGGCGGCCUGGAACUGCAUCUAAGGAGUUCGGAAGACGGAGACAACGAUCUUGUUCAGUUUUUGAUGGUGCAAGAAAAGAGCUCCAUCAAAAUCA (((((............(((.(((((((.......)))).))).)))...((....)).)))))..........((((..((((((......))))))..))))............. (-16.87 = -16.40 + -0.47)

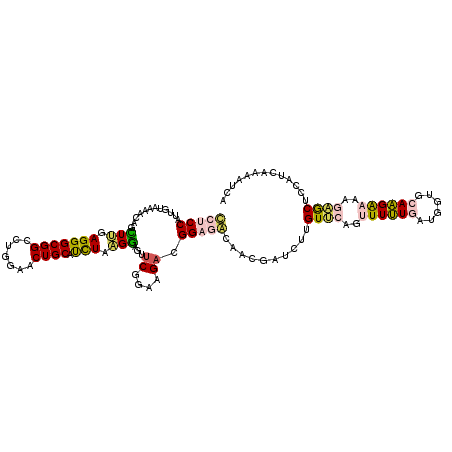
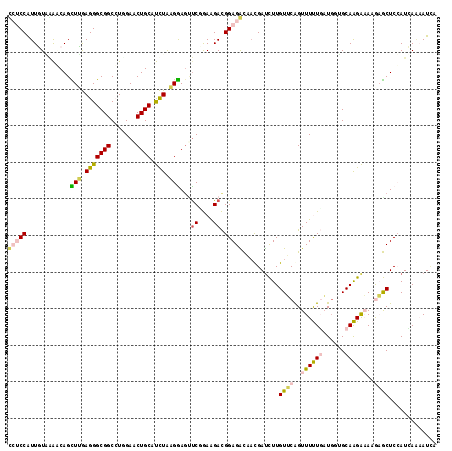
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:53:40 2006