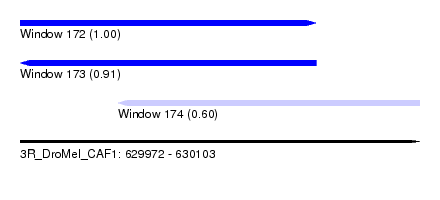
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 629,972 – 630,103 |
| Length | 131 |
| Max. P | 0.996107 |

| Location | 629,972 – 630,069 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -20.91 |
| Energy contribution | -22.72 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 629972 97 + 27905053 AAAUCGUUUAGGUAUGCUCGUCUUGAAUACG--------UGUAUGAAUGUAGAACCUAUGAUUUAGAGCACAUUUGCCGAAAUCGGCAAGUGCCU-CACCACGAUA ..(((((...(((.((((((((.....((((--------(......)))))........)))...)))))(((((((((....)))))))))...-.)))))))). ( -29.42) >DroSec_CAF1 45751 96 + 1 AAAUCGUUUAGGAAUGCUCGUCUUGAAUUCA--------UGUACGAAUGUAGAUCCUAUCAUUUAGAGCACAUUUGCCGAAAUCGGCAAGUGCCUACACCACGA-- ...((((.((((..(((((....(((....(--------(.(((....))).))....)))....)))))(((((((((....)))))))))))))....))))-- ( -29.50) >DroSim_CAF1 47377 98 + 1 AAAUCGGUUAGGAAUGCUCGUCUUGAAUUCA--------UGUACGAAUGUAGAUCCUAUCAUUUAGAGCACAUUUGCCGAAAUCGGCAAGUGCCUACACCACGAUA .....(((((((..(((((....(((....(--------(.(((....))).))....)))....)))))(((((((((....))))))))))))).)))...... ( -32.00) >DroEre_CAF1 49173 106 + 1 AAAUCGUUUAGAAUCGUUCAGAUGUAAUACGAAUGGUUUUGUACGAAUGAAGAUAGUAUCGUUUAGAGAACAUUUGCCGAAAUCGGCAAAUGCCUACGCCACGAUA ...((((.(((((((((((...........))))))))))).))))..........((((((...(((..(((((((((....))))))))).)).)...)))))) ( -32.20) >consensus AAAUCGUUUAGGAAUGCUCGUCUUGAAUACA________UGUACGAAUGUAGAUCCUAUCAUUUAGAGCACAUUUGCCGAAAUCGGCAAGUGCCUACACCACGAUA ..(((((.((((..(((((........((((((....)))))).(((((..........))))).)))))(((((((((....)))))))))))))....))))). (-20.91 = -22.72 + 1.81)

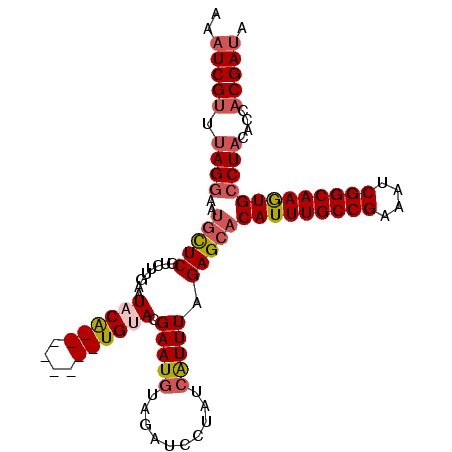
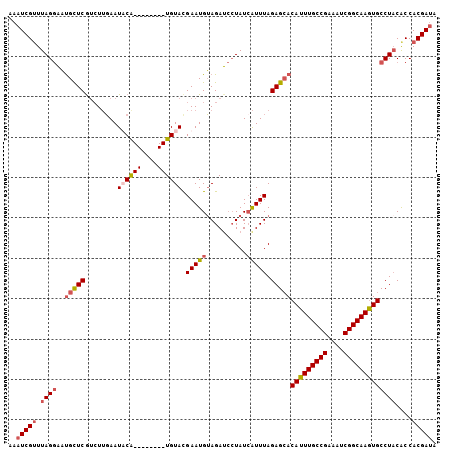
| Location | 629,972 – 630,069 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 629972 97 - 27905053 UAUCGUGGUG-AGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUCAUAGGUUCUACAUUCAUACA--------CGUAUUCAAGACGAGCAUACCUAAACGAUUU .(((((....-(((...((((((....)))))).(((((((......(((.((.............)--------).))).......))))))))))..))))).. ( -24.04) >DroSec_CAF1 45751 96 - 1 --UCGUGGUGUAGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUGAUAGGAUCUACAUUCGUACA--------UGAAUUCAAGACGAGCAUUCCUAAACGAUUU --((((....(((((((((((((....))))))..)))).))).....(((((......(((((.(.--------((....)).))))))...))))).))))... ( -28.20) >DroSim_CAF1 47377 98 - 1 UAUCGUGGUGUAGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUGAUAGGAUCUACAUUCGUACA--------UGAAUUCAAGACGAGCAUUCCUAACCGAUUU .....((((.(((((((((((((....))))))..)))).))).....(((((......(((((.(.--------((....)).))))))...))))))))).... ( -28.50) >DroEre_CAF1 49173 106 - 1 UAUCGUGGCGUAGGCAUUUGCCGAUUUCGGCAAAUGUUCUCUAAACGAUACUAUCUUCAUUCGUACAAAACCAUUCGUAUUACAUCUGAACGAUUCUAAACGAUUU ((((((...(..(((((((((((....)))))))))))..)...))))))........................((((.....(((.....))).....))))... ( -25.50) >consensus UAUCGUGGUGUAGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUGAUAGGAUCUACAUUCGUACA________CGAAUUCAAGACGAGCAUUCCUAAACGAUUU ..((((....(((((((((((((....))))))..)))).))).....(((((......(((((.....................)))))...))))).))))... (-17.48 = -18.35 + 0.87)

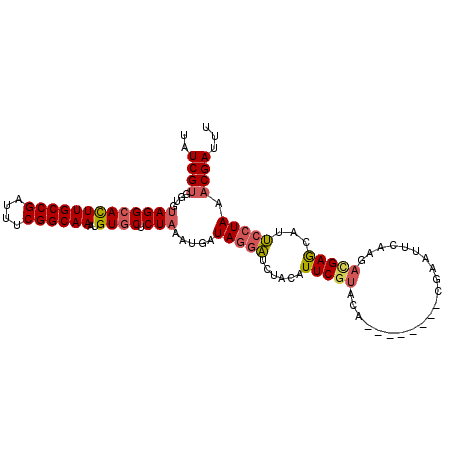
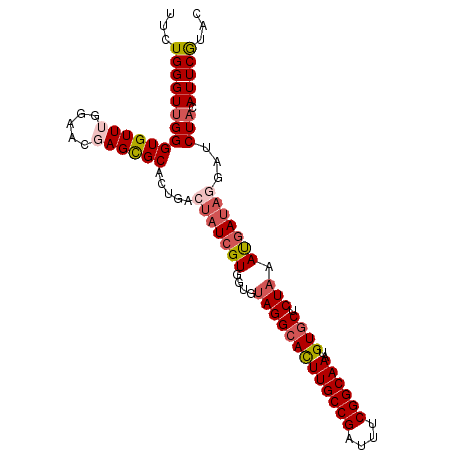
| Location | 630,004 – 630,103 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -22.17 |
| Energy contribution | -23.93 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 630004 99 - 27905053 UUCUGGGUUGGGUGUUUGGAACGAGUGCACUAACUAUCGUGGUG-AGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUCAUAGGUUCUACAUUCAUAC ........(((((((..(((((.(((......)))...(((((.-.(((((((((((....))))))..)))))....)))))..))))))))))))... ( -30.60) >DroSec_CAF1 45783 93 - 1 UUCUGGGUUGGGUGUUUGGAACUAGCGCA-------UCGUGGUGUAGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUGAUAGGAUCUACAUUCGUAC ...((.(((((..........))))).))-------.((.(((((((((((((((((....))))))..))))((((.....))))..)))))))))... ( -27.50) >DroSim_CAF1 47409 100 - 1 UUCUGGGUUGGGUGUUUGGAACUAGCGCACUGACUAUCGUGGUGUAGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUGAUAGGAUCUACAUUCGUAC ...(((((((((((((.......))))).))..(((((((....(((((((((((((....))))))..)))).))).)))))))))))))......... ( -32.20) >DroEre_CAF1 49213 100 - 1 UUCUGGGUUGGGUGUUUGGAACGAGCGCACUGACUAUCGUGGCGUAGGCAUUUGCCGAUUUCGGCAAAUGUUCUCUAAACGAUACUAUCUUCAUUCGUAC .....(((..((((((((...))))))).)..)))(((((...(..(((((((((((....)))))))))))..)...)))))................. ( -33.80) >consensus UUCUGGGUUGGGUGUUUGGAACGAGCGCACUGACUAUCGUGGUGUAGGCACUUGCCGAUUUCGGCAAAUGUGCUCUAAAUGAUAGGAUCUACAUUCGUAC ...((((((((((((((.....)))))).....(((((((....(((((((((((((....))))))..)))).))).)))))))...))).)))))... (-22.17 = -23.93 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:18 2006