| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,601,334 – 27,601,435 |
| Length | 101 |
| Max. P | 0.610080 |

| Location | 27,601,334 – 27,601,435 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.66 |
| Mean single sequence MFE | -17.44 |
| Consensus MFE | -10.18 |
| Energy contribution | -10.27 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27601334 101 - 27905053 UGAGAUCUUUU------CUUUUUCAGAUUACUCCUUUCCGUACCUUAGCGUGGCCAUAUCCAUGGUAGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCAAACCC .((((.....)------))).(((((.............((......))((((........((((((.....))))))))))............)))))........ ( -14.40) >DroVir_CAF1 3022 99 - 1 UGACU-UA-AU------UUUUUUUAGAUUAUUCGUUUCCUUAUCUAAGUAUCGCUAUGUCAAUGGUGGCCAACGCCAUUCACUACUCUUUAAAGCUAGAUCAAACCC .(((.-..-..------....(((((((.............))))))).........)))(((((((.....)))))))..(((((......)).)))......... ( -14.59) >DroPse_CAF1 4704 100 - 1 AAAUAUCAU-U------GUCUUACAGACUACUCCUUUCCGUAUCUUAGCAUGGCCAUUUCUAUGGUGGCCAAUGCCAUACACUAUUCCCUCAAACUGGACCAAACCC .........-.------(((.....)))...........((((....(((((((((((.....))))))).)))).)))).....(((........)))........ ( -19.70) >DroWil_CAF1 3671 107 - 1 UUAUAUCAUUUACUGUGAUCUUUCAGAUUAUUCGUUUCCCUAUCUCAGUGUGGCCAUAUCGAUGGUCGCCAACGCCAUUCACUAUUCACUGAAAAUAGAUCAAACAC ..............((((((.....))))))..((((..((((.((((((.((((((....))))))((....))...........))))))..))))...)))).. ( -20.80) >DroYak_CAF1 3150 101 - 1 UGACAUGUAUA------CCUUUUCAGAUUACUCUUUUCCCUACCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCAAACCC ...........------..............................((((((((((.......)))))).)))).(((((..............)))))....... ( -15.84) >DroPer_CAF1 4715 100 - 1 AAAUAUCAU-U------GUCUUACAGACUACUCCUUUCCGUAUCUUAGCGUGGCCAUUUCUAUGGUGGCCAAUGCCAUACACUAUUCCCUCAAACUGGACCAAACCC .........-.------(((.....)))...........((((....(((((((((((.....))))))).)))).)))).....(((........)))........ ( -19.30) >consensus UGAUAUCAUUU______CUCUUUCAGAUUACUCCUUUCCGUAUCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUACACUAUUCCCUCAAACUGGAUCAAACCC ...............................................((((((((((.......)))))).))))..........(((........)))........ (-10.18 = -10.27 + 0.09)


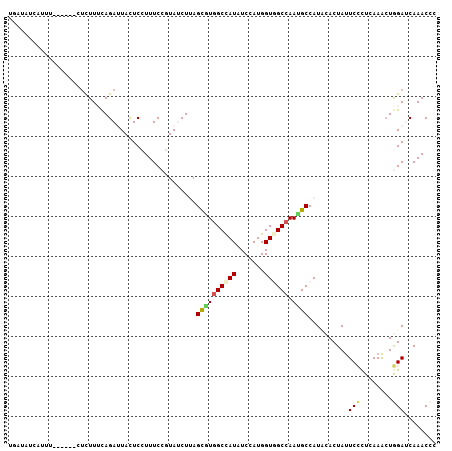
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:53:28 2006