
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,563,761 – 27,563,966 |
| Length | 205 |
| Max. P | 0.969736 |

| Location | 27,563,761 – 27,563,863 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 59.69 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
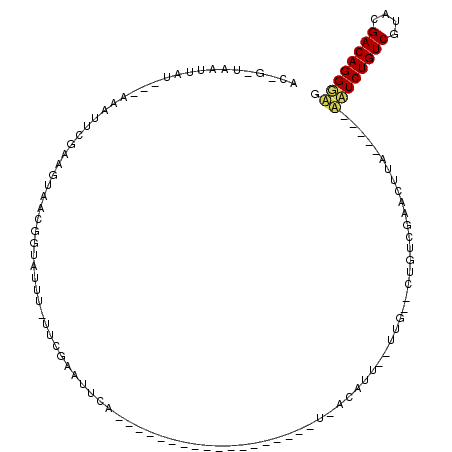
>3R_DroMel_CAF1 27563761 102 + 27905053 ACUGUUAAUUACAUUACAUUACUAUUAACAGUAUUUUUUCAAAUUCC--------------UUUCUAACAUUU-UUGUUCUGUCAAAUUUUCUAUUGUUCUGUCGUACGACAGGACG (((((((((..............)))))))))...............--------------.....((((...-.)))).................((((((((....)))))))). ( -19.34) >DroEre_CAF1 85933 103 + 1 AC---UAAUCAU---AAAUUCGAAGUAACGGUAUUU-UUCGAAUUCAUCAGCGGGACAACCAACUUUACAUUAAUUG--CUAUCGAACUUC-----AAUCUGUCGUACGACAGGGAG ..---...((..---...(((((((((((((.....-.)))...........((.....)).............)))--)).)))))....-----..((((((....)))))))). ( -18.40) >DroYak_CAF1 84229 79 + 1 ACCGAUAAUUAU---AAAGUCGAAGUAACGGUAUUU-UUCGAAUUCA---------------------------UUG--CUGUCGAACUUA-----ACUCUGUCGUACGACAGGGUG ..(((((.....---....((((((..........)-))))).....---------------------------...--.)))))......-----((((((((....)))))))). ( -19.97) >consensus AC_G_UAAUUAU___AAAUUCGAAGUAACGGUAUUU_UUCGAAUUCA__________________U_ACAUU__UUG__CUGUCGAACUUA_____AAUCUGUCGUACGACAGGGAG ................................................................................................((((((((....)))))))). ( -9.54 = -9.77 + 0.23)


| Location | 27,563,823 – 27,563,938 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -19.43 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
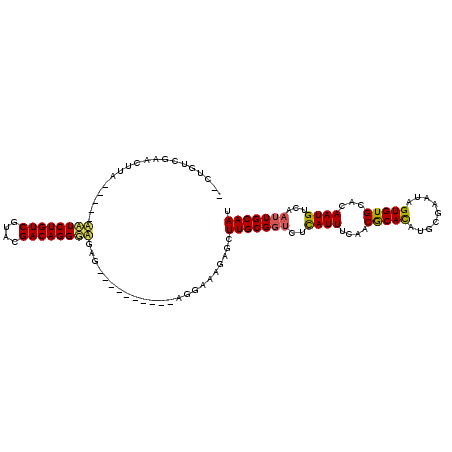
>3R_DroMel_CAF1 27563823 115 + 27905053 UUCUGUCAAAUUUUCUAUUGUUCUGUCGUACGACAGGACGAGAGAGAGAGAAAGGAAGGAGCUUGCG-UCUUAUUUCAACGCAUAAGCGAAUAGUGUGCACAAUGUCUUUUGCAAU ((((.((...((((((.((((((((((....))))))))))...))))))....)).))))..((((-(.........)))))...(((((..(..(.....)..)..)))))... ( -31.30) >DroEre_CAF1 86003 98 + 1 --CUAUCGAACUUC-----AAUCUGUCGUACGACAGGGAG-U----------AGAAAAGAGCUUGCGGUCUCAUUUCAACACACUUUCGAAUAGUGUGCACAAUGUCAAUUGCAAU --...((..(((((-----...(((((....)))))))))-)----------.)).......(((((((..((((....((((((.......))))))...))))...))))))). ( -24.20) >DroYak_CAF1 84275 99 + 1 --CUGUCGAACUUA-----ACUCUGUCGUACGACAGGGUGAG----------AGGAAAGAGCUUGCGGUCUCAUUUCAACGCACAUGCGAAUAGUGUGCAAAAUGUCAAUUGCAAU --...((...(((.-----((((((((....))))))))..)----------))....))..(((((((..(((((....((((((.......)))))).)))))...))))))). ( -29.00) >consensus __CUGUCGAACUUA_____AAUCUGUCGUACGACAGGGAGAG__________AGGAAAGAGCUUGCGGUCUCAUUUCAACGCACAUGCGAAUAGUGUGCACAAUGUCAAUUGCAAU ...................((((((((....)))))))).......................(((((((..((((....(((((.........)))))...))))...))))))). (-19.43 = -19.67 + 0.23)

| Location | 27,563,863 – 27,563,966 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27563863 103 + 27905053 AGAGAGAGAGAAAGGAAGGAGCUUGCG-UCUUAUUUCAACGCAUAAGCGAAUAGUGUGCACAAUGUCUUUUGCAAUUAGUGGUGAAUGUGCAUACUUUUAGUGA .((((..((((.(((......)))...-)))).))))..(((....)))((.((((((((((...((..(..(.....)..).)).)))))))))).))..... ( -25.30) >DroEre_CAF1 86036 93 + 1 -U----------AGAAAAGAGCUUGCGGUCUCAUUUCAACACACUUUCGAAUAGUGUGCACAAUGUCAAUUGCAAUUAGAGGUGAGUGUGCAUACUUGUAUUGU -.----------......(((..((((..((((((((..((((((.......))))))..((((....))))......))))))))..))))..)))....... ( -24.70) >DroYak_CAF1 84308 92 + 1 AG----------AGGAAAGAGCUUGCGGUCUCAUUUCAACGCACAUGCGAAUAGUGUGCAAAAUGUCAAUUGCAAUUAGAGGUGAGU--GCAUAAUUUUACUGG ..----------.((.((((...((((..((((((((..(((....)))..((((.(((((........))))))))))))))))))--)))...)))).)).. ( -22.90) >consensus AG__________AGGAAAGAGCUUGCGGUCUCAUUUCAACGCACAUGCGAAUAGUGUGCACAAUGUCAAUUGCAAUUAGAGGUGAGUGUGCAUACUUUUACUGA ................(((((..(((...((((((((..(((((.........))))).....(((.....)))....))))))))...)))..)))))..... (-15.50 = -16.07 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:53:14 2006