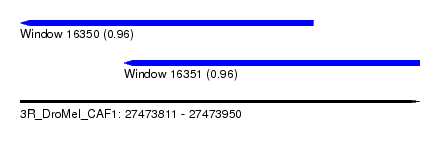
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,473,811 – 27,473,950 |
| Length | 139 |
| Max. P | 0.959014 |

| Location | 27,473,811 – 27,473,913 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.61 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27473811 102 - 27905053 GGCGCGCAUUUUCAACAGCUGAAGCUAA------------AGCUGCAGCUGAAAAGCUGAAGUGGAAAGCUGCACUGGGAGAAAUUAAACAGCCAUUCGAGG-UUUUAUCAAACA--- (((((((.((((((.(((((........------------)))))(((((....)))))...)))))))).))..((.((....))...))))).......(-(((....)))).--- ( -29.00) >DroSec_CAF1 29625 106 - 1 GGCGCGCAUUUUGAACAGCUGAAGCUGA------------AACUCCAGCUGAAAAGCUGAAGUGGAAAGCUGCACUGGGAGAAAUUGCACAGCCAUUUGAAGAUUUAAUCAAUGCUUU ((((.(((((((...(((.((.((((..------------.(((.(((((....))))).)))....)))).)))))...)))).))).).)))..((((........))))...... ( -32.70) >DroSim_CAF1 29552 106 - 1 GGCGCGCAUUUUCAACAGCUGAAGCUGA------------AGCUGCAGCUGAAAAGCUGAAGUGGAAAGCUGCACUGGGAGAAAUUGCACAGCCAUUCGAGGAUUUUAUCAAACCUUU ((((.(((.((((..(((.((.((((..------------.(((.(((((....))))).)))....)))).)))))...)))).))).).)))....((((.(((....))))))). ( -35.80) >DroEre_CAF1 29562 94 - 1 GGCGCGCAUUUUCAACAGCUGAAG------------------CUGCAGCUGAAAAGCUGAAGUGGAAAGCUGCACCGCGGAAAACUGAAUGGCCAUUCGAGGAUUUCAUCCA------ ((.((((.((((((.((((....)------------------)))(((((....)))))...)))))))).)).))..(((....((((...((......))..))))))).------ ( -31.30) >DroYak_CAF1 30818 100 - 1 GGCGCGCAUUUUCAACAGCUGAAG------------------CUGCAGCUGAAAAGCUGAAGUGGAAAGCUGCACUGGCAAAAACUGUAUGGCCAUUUGAAGAUUUCAUCAGACUGUU (((((((.((((((.((((....)------------------)))(((((....)))))...)))))))).))....(((.....)))...))).(((((.(....).)))))..... ( -28.90) >DroAna_CAF1 43238 114 - 1 GGCGCGCAAUUUCAACAGCUGAAUCUGAAACUGAAUCUGAAGCUGAGGCUGAAAAGCCAAAGUGGGUGGCUAUACACUGAAAAACA----AGCUCUCGGAAUAAUUAAUUAGACUAAC (((..............)))...(((((.......(((((((((..((((....))))....(.((((......)))).)......----)))).)))))........)))))..... ( -24.50) >consensus GGCGCGCAUUUUCAACAGCUGAAGCUGA____________AGCUGCAGCUGAAAAGCUGAAGUGGAAAGCUGCACUGGGAAAAACUGAACAGCCAUUCGAAGAUUUAAUCAAACC_UU (((............((((((.((((..............)))).))))))...((((.........))))....................)))........................ (-17.49 = -17.61 + 0.11)



| Location | 27,473,847 – 27,473,950 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -19.61 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27473847 103 - 27905053 GAGCGACAAACUUGCAAAAGUU---UUGUUGGUGGCCAGCGGCGCGCAUUUUCAACAGCUGA-AGCUAA------------AGCUGCAGCUGAAAAGCUGAA-GUGGAAAGCUGCACUGG .(.((((((((((....))).)---)))))).)..((((..(((.((.((((((.(((((..-......------------)))))(((((....)))))..-.))))))))))).)))) ( -35.80) >DroPse_CAF1 35034 106 - 1 GAGCGAGAAACUUUCAAAAGUUUUUUUGUUGGUCGCCAGCGGCGCGCAUUUUCAACAGCUGAGAGCUG-------------AGCUGAAGCUGAAAAGCUGGAAGCUGGUAGCCAGAG-CU .((((((((((((....)))))))))..(((((..(((((..(............(((((.(....).-------------))))).((((....)))).)..)))))..))))).)-)) ( -39.20) >DroSec_CAF1 29665 103 - 1 GAGCGACAAACUUGCAAAAGUU---UUGUUGGUCGCCAGCGGCGCGCAUUUUGAACAGCUGA-AGCUGA------------AACUCCAGCUGAAAAGCUGAA-GUGGAAAGCUGCACUGG ((.((((((((((....))).)---)))))).)).((((..(((.((.((((...((((...-.)))).------------.(((.(((((....))))).)-)).))))))))).)))) ( -37.10) >DroEre_CAF1 29596 97 - 1 GAGCGACAAACUUGCAAAAGUU---UUGUUGGUCGCCAGCGGCGCGCAUUUUCAACAGCUGA-AG------------------CUGCAGCUGAAAAGCUGAA-GUGGAAAGCUGCACCGC ((.((((((((((....))).)---)))))).))....((((.((((.((((((.((((...-.)------------------)))(((((....)))))..-.)))))))).)).)))) ( -40.00) >DroAna_CAF1 43274 115 - 1 AAGCGACAAACUUGAUAAAGUU---UUGUUGGUCGCCAGCGGCGCGCAAUUUCAACAGCUGA-AUCUGAAACUGAAUCUGAAGCUGAGGCUGAAAAGCCAAA-GUGGGUGGCUAUACACU ..(((((((((((....)))))---).....))))).(((.((.(((........(((((((-.((.......)).))...))))).((((....))))...-))).)).)))....... ( -36.90) >DroPer_CAF1 32901 106 - 1 GAGCGAGAAACUUUCAAAAGUUUUUUUGUUGGUCGCCAGCGGCGCGCAUUUUCAACAGCUGAGAGCUG-------------AGCUGAAGCUGAAAAGCUGGAAGCUGGUAGCCAGAG-CU .((((((((((((....)))))))))..(((((..(((((..(............(((((.(....).-------------))))).((((....)))).)..)))))..))))).)-)) ( -39.20) >consensus GAGCGACAAACUUGCAAAAGUU___UUGUUGGUCGCCAGCGGCGCGCAUUUUCAACAGCUGA_AGCUG_____________AGCUGAAGCUGAAAAGCUGAA_GUGGAAAGCUACAC_CU ..(((...(((((....)))))...(((((((...))))))))))((........((((.....))))...................((((....))))...........))........ (-19.61 = -20.17 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:51 2006