
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
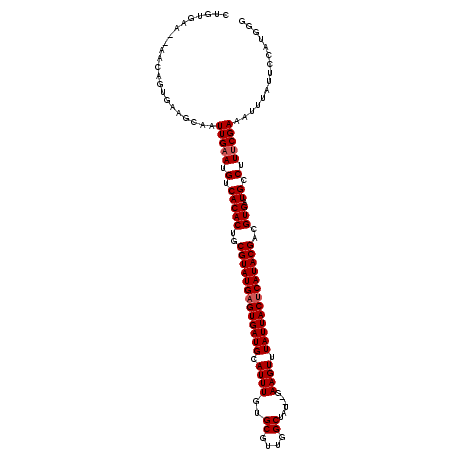
| Location | 27,468,416 – 27,468,533 |
| Length | 117 |
| Max. P | 0.867951 |

| Location | 27,468,416 – 27,468,514 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27468416 98 + 27905053 ----------------------UUGAAUGUCACACUGCGUAUGAGUGAUGCAUUUGUGCGUUGGCUAUGCGAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGG ----------------------(((((.(.(((((..(((((((((((((.((((.(((((.....))))))))).)))))))))))))..))).)).).)))))............... ( -30.80) >DroSec_CAF1 24280 118 + 1 CUGUGAAAAAACAGUGAAGCAAUUGAAUGUCACACUGCGUAUGAGUGAUGAAUUUCUGCGUUGGCUAU--GAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGG ((((......))))........(((((.(.(((((..(((((((((((((((((((.((....))...--)))))))))))))))))))..))).)).).)))))............... ( -38.20) >DroSim_CAF1 24169 118 + 1 CUGUGAAAAAACAGUGAAGCAAUUGUAUGUCACACUGCGUAUGAGUGAUGCAUUUCUGCGUUGGCUAU--GAAGUUUAUUACGCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGG (((((.....(((((......)))))..(((((....((((((.((((((.(((((.((....))...--))))).)))))).))))))..)))))..................))))). ( -30.30) >DroEre_CAF1 24222 116 + 1 CUAUGAA--AACAGUGUAGCAAUUGAAUGUCACACUGCGUAUGAGUGAUGCAUUUGCGCGAUGGCUGU--GAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUGUAUUUCAUGGG (((((((--(.((((((..((......))..))))))(((((((((((((..((..(((....)).).--.))...)))))))))))))......................)))))))). ( -39.10) >DroYak_CAF1 25336 116 + 1 CUGUGAA--AACAGUGAAGCAAUUGAAUGUCACACUGCGUAUGAGUGAUGCAUUUGUGCGUUGGCUGU--GAAGUUUAUUACUCAUACGACGUGUUGCCCUUCGAAAUUUAUUUCAUGGG (((((((--....(((((....(((((.((.((((..(((((((((((((..((..(((....)).).--.))...)))))))))))))..)))).))..)))))..)))))))))))). ( -34.40) >consensus CUGUGAA__AACAGUGAAGCAAUUGAAUGUCACACUGCGUAUGAGUGAUGCAUUUGUGCGUUGGCUAU__GAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGG ......................(((((.(.(((((..(((((((((((((.((((..((....))......)))).)))))))))))))..))).)).).)))))............... (-23.74 = -24.14 + 0.40)

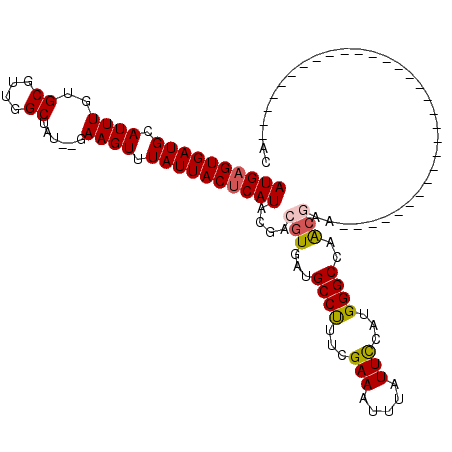
| Location | 27,468,434 – 27,468,533 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27468434 99 + 27905053 AUGAGUGAUGCAUUUGUGCGUUGGCUAUGCGAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGGCUAACGAACUCUUCCG---------------------CAC ((((((((((.((((.(((((.....))))))))).))))))))))....(((...((((...(((.....)))...))))..)))..........---------------------... ( -25.80) >DroSec_CAF1 24320 86 + 1 AUGAGUGAUGAAUUUCUGCGUUGGCUAU--GAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGGCCAACAAA-------------------------------- ((((((((((((((((.((....))...--)))))))))))))))).....((...((((...(((.....)))...))))..))...-------------------------------- ( -27.20) >DroSim_CAF1 24209 86 + 1 AUGAGUGAUGCAUUUCUGCGUUGGCUAU--GAAGUUUAUUACGCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGGCCAACGAA-------------------------------- .........(((....)))(((((((..--((((..((((((((....).)))))))..))))(((.....)))....)))))))...-------------------------------- ( -20.90) >DroEre_CAF1 24260 118 + 1 AUGAGUGAUGCAUUUGCGCGAUGGCUGU--GAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUGUAUUUCAUGGGCCAGCGAACUCGUGCGUAUCUGUAAUGCUUUAUCAAGAAC ..(((((.((((..(((((..(((((..--((((..((((((((....)).))))))..))))((((....))))...)))))(((....))))))))..)))).))))).......... ( -31.20) >DroYak_CAF1 25374 97 + 1 AUGAGUGAUGCAUUUGUGCGUUGGCUGU--GAAGUUUAUUACUCAUACGACGUGUUGCCCUUCGAAAUUUAUUUCAUGGGCCAA----------------UGUAACG-----UCAAGAAC ((((((((((..((..(((....)).).--.))...))))))))))..((((((((((((...((((....))))..))).)))----------------)...)))-----))...... ( -27.20) >consensus AUGAGUGAUGCAUUUGUGCGUUGGCUAU__GAAGUUUAUUACUCAUACGACGUGAUGCCUUUCGAAAUUUAUUCCAUGGGCCAACGAA______________________________AC ((((((((((.((((..((....))......)))).))))))))))....(((...((((...(((.....)))...))))..))).................................. (-16.24 = -16.48 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:47 2006