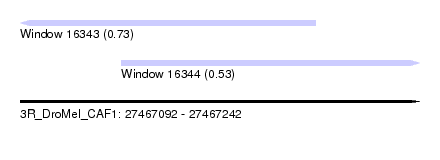
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,467,092 – 27,467,242 |
| Length | 150 |
| Max. P | 0.732598 |

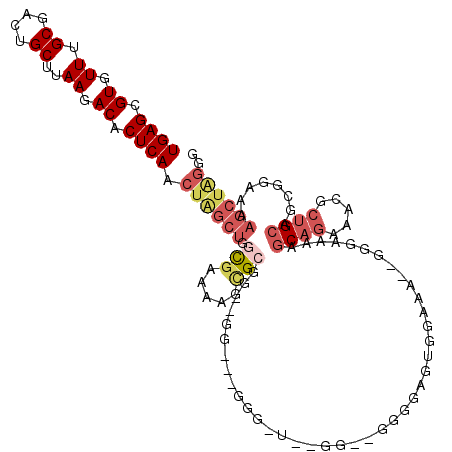
| Location | 27,467,092 – 27,467,203 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -14.73 |
| Energy contribution | -16.15 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732598 |
| Prediction | RNA |
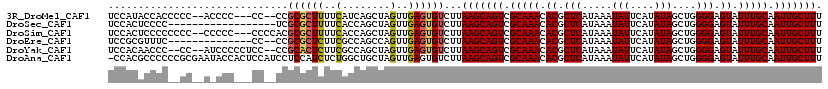
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27467092 111 - 27905053 UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUAGCUGAUGAAAAGCGCGG--GG---GGGGU--GGGGGUGGUAUGGAAG--GGGGAAAGCAGAAACGCUGCAGCGGAAAAGCUAGCG ..(((((.(((((.((..(((....(((((..((.(((.......)))...)--).---.))))--).)))..)).......--.......))))).)))))(((((......))).)). ( -36.46) >DroSec_CAF1 22980 102 - 1 UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUAGCUGGUGAAAAGCGCGA------------------GGGGAGUGGAAAGCGGGAAAAGCAGAAACGCUGCAGCGGAAAAGCUAGGG ..(((((.(((((..((((((....(((((..((.(((.......)))...)------------------)..)))))..)))))).....))))).)))))..(((......))).... ( -37.30) >DroSim_CAF1 22844 115 - 1 UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUAGCUGGUGAAAAGCGCGUGGGG---GGGGG--GGGGGGGGAGUGGAAAGCGGGAAAAGCAGAAACGCUGCAGCGGAAAAGCUAGGG ..(((((.(((((..((((((....(((((..((.(((.......))).(......---)....--...))..)))))..)))))).....))))).)))))..(((......))).... ( -34.10) >DroEre_CAF1 22863 104 - 1 UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUGGCUGGCGAAGAGCGCGG--GG--------------GAAACGCGGAAAAGCGGAAAAGCAGAAACGCUGCAGCGGAAAAGCUGGGG ..(((((.(((((..((((((..(.....)..(((.((.(((.....))).)--)(--------------....).)))..))))))....))))).))))).((((......))))... ( -34.20) >DroYak_CAF1 23912 114 - 1 UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUAGCUGGCGAAGAGUGCGG--GGAGGGGGAU--GG--GGGUUGUGGAAAAGUGGAAAAGCAGAAACGCAGCAGCGGAAAAGCUAGGG ...((((.(((((..(..(((..(((.((((.((..((.(((.....))).)--)...))...)--))--).)))......)))..)....))))).))))...(((......))).... ( -28.60) >DroAna_CAF1 35302 99 - 1 UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUAGCAGCCAGAGAUGGAGGAUGGAGUGGUAUUCGCGGGGGGCGUGG-------------CAGGUUGAGUGCAACGGAAG-------- ((((((.(((...))))))))).(.(((((((((.((.(((.....(.(.(((((......))))).).)..)))...)-------------).))))))))))..(....)-------- ( -30.00) >consensus UGAGCGUGUUUGCGACUGCUUAAGACACUCAACUAGCUGGCGAAAAGCGCGG__GG___GGG_U__GG__GGGGAGUGGAAA__GGGAAAAGCAGAAACGCUGCAGCGGAAAAGCUAGGG ((((.((.((.((....))..)).)).)))).((((((.(((.....))).........................................((((.....))))........)))))).. (-14.73 = -16.15 + 1.42)


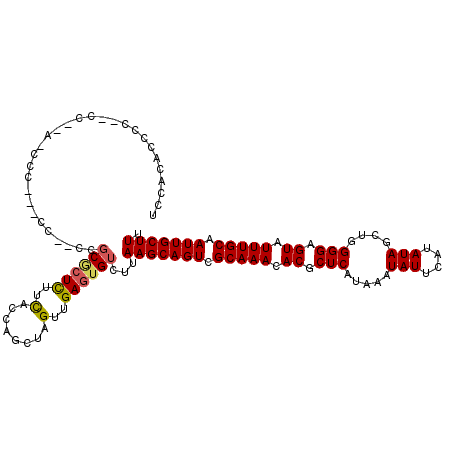
| Location | 27,467,130 – 27,467,242 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27467130 112 + 27905053 UCCAUACCACCCCC--ACCCC---CC--CCGCGCUUUUCAUCAGCUAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU ..............--....(---((--(.(((((.......)))....(((((((.....((....))....)))))))..............)).))))((((........)))).. ( -22.60) >DroSec_CAF1 23020 101 + 1 UCCACUCCCC------------------UCGCGCUUUUCACCAGCUAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU ...(((((((------------------..(((((.......)))....(((((((.....((....))....)))))))..............)).)))))))....((....))... ( -26.80) >DroSim_CAF1 22884 114 + 1 UCCACUCCCCCCCC--CCCCC---CCCCACGCGCUUUUCACCAGCUAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU ...(((((((....--.....---........((((..((((((....))).)))....))))............(((.(((........)))))).)))))))....((....))... ( -24.20) >DroEre_CAF1 22903 103 + 1 UCCGCGUUUC--------------CC--CCGCGCUCUUCGCCAGCCAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU ...(((.(((--------------((--(.(((((.......)))....(((((((.....((....))....)))))))..............)).))))))....)))......... ( -26.20) >DroYak_CAF1 23952 113 + 1 UCCACAACCC--CC--AUCCCCCUCC--CCGCACUCUUCGCCAGCUAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU ..........--..--..........--..(((..((((.(((((((..(((((((.(((.(.((.((......))))).))))))))))..)))))))))))....)))......... ( -26.50) >DroAna_CAF1 35322 118 + 1 -CCACGCCCCCCGCGAAUACCACUCCAUCCUCCAUCUCUGGCUGCUAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU -...........((((((((..(((((....(((....)))..((((..(((((((.(((.(.((.((......))))).))))))))))..))))))))).))))))))......... ( -28.00) >consensus UCCACACCCC__CC__A_CCC___CC__CCGCGCUCUUCACCAGCUAGUUGAGUGUCUUAAGCAGUCGCAAACACGCUCAUAAAUAUUCAUAUAGCUGGGGAGUAUUUGCAAUUGCUUU ..............................((((((..(........)..))))))...(((((((.(((((.((.(((.....(((....)))....))).)).))))).))))))). (-21.25 = -21.25 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:44 2006