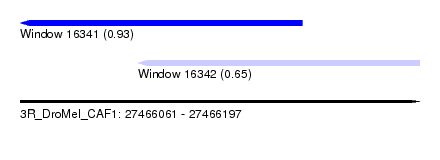
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,466,061 – 27,466,197 |
| Length | 136 |
| Max. P | 0.925944 |

| Location | 27,466,061 – 27,466,157 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -21.62 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
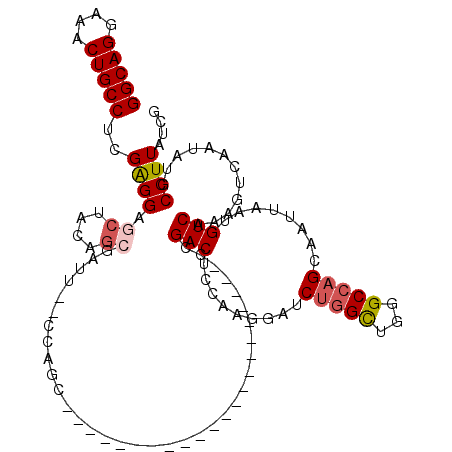
>3R_DroMel_CAF1 27466061 96 - 27905053 GGCAGGAAACUGCCUCGAGGAGCUAUAGCGAUU--CCAGC----------------------CCGACUCCAAGGAUCUGGCUGGGCCAGCAAUUAAUGUCAAAGUCAAUAUGCCUUAUCG (((((....))))).(((.(((.((((((...(--(((((----------------------(.((.((....)))).)))))))...))......((.......)).)))).))).))) ( -33.50) >DroSec_CAF1 21961 96 - 1 GGCAGGAAACUGCCUCGAGGAGCUACAGCGAUU--CCAGC----------------------CCGACUCCAAGGAUCUGGCUGGGCCAGCAAUUAAUGUCAAAGUCAACAUGCCUUAUCG (((((....)))))..((((.......((...(--(((((----------------------(.((.((....)))).)))))))...)).....((((........)))).)))).... ( -34.70) >DroSim_CAF1 21832 96 - 1 GGCAGGAAACUGCCUCGAGGAGCUACAGCGAUU--CCAGC----------------------CCGACUCCAAGGAUCUGGCUGGGCCAGCAAUUAAUGUCAAAGUCAAUAUGCCUUAUCG (((((....)))))..((((.......((...(--(((((----------------------(.((.((....)))).)))))))...)).....((((........)))).)))).... ( -32.80) >DroEre_CAF1 21800 110 - 1 GGCAGGAAACUGCCUCGAGGAGCUCGAGCAGCU--CGAGGAGCUCGAGGAGCUACAG------CGACUCCA--GAUCUGGCUGGGCCAGCAAUUAAUGUCAAAGUCAAUAUGCCUUAUCG (((((....)))))..(((..(((..(((..((--((((...))))))..)))..))------)..)))..--(((..(((..(((..(((.....)))....))).....)))..))). ( -41.10) >DroYak_CAF1 22831 109 - 1 GGCAGGAAACUGCCUCGAGGAGCUCGAGAAGCU-----------CGAGAAGCUACAGAGAUUCCGACUCCAAGGAUCUGGCUGGGCCAGCAAUUAAUGUCAAAGUCAAUAUGCCUUAUCG (((((....))))).(((((((.(((.(((.((-----------(...........))).))))))))))((((...(((((..(.((........)).)..))))).....)))).))) ( -36.80) >DroPer_CAF1 25394 94 - 1 GGCAGGAAACUGCCUCGGGGCUCCGUCUCUCUUUCUCUAU----------------------CCGACUCC----CUCUAGUUGGGCCCGCAAUUAAUGUCGAAGUCAAUAUGCCUUAUCG (((((....)))))..(((((..................(----------------------((((((..----....)))))))..((((.....)).))..........))))).... ( -25.20) >consensus GGCAGGAAACUGCCUCGAGGAGCUACAGCGAUU__CCAGC______________________CCGACUCCAAGGAUCUGGCUGGGCCAGCAAUUAAUGUCAAAGUCAAUAUGCCUUAUCG (((((....)))))..((((.((....))...................................(((.........(((((...)))))........)))............)))).... (-21.62 = -22.18 + 0.56)


| Location | 27,466,101 – 27,466,197 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -19.55 |
| Energy contribution | -20.61 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27466101 96 - 27905053 UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGAGCUAUAGCGAUUCCAGC----------------------CCGACUCCAAGGAUCUGG .((((.(((((.....(((.........))).((((..(.(((((....))))).)...))))..)))))....))))----------------------(((..((....))..))) ( -31.40) >DroSec_CAF1 22001 96 - 1 UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGAGCUACAGCGAUUCCAGC----------------------CCGACUCCAAGGAUCUGG .((((((((...................))))((((..(.(((((....))))).)...))))..))))....((((.----------------------((........))..)))) ( -32.91) >DroSim_CAF1 21872 96 - 1 UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGAGCUACAGCGAUUCCAGC----------------------CCGACUCCAAGGAUCUGG .((((((((...................))))((((..(.(((((....))))).)...))))..))))....((((.----------------------((........))..)))) ( -32.91) >DroEre_CAF1 21840 110 - 1 UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGAGCUCGAGCAGCUCGAGGAGCUCGAGGAGCUACAG------CGACUCCA--GAUCUGG .((((........................((.((((..(.(((((....))))).)...)))).))(((..((((((...))))))..))).)))------)....(((--....))) ( -41.00) >DroYak_CAF1 22871 109 - 1 UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGAGCUCGAGAAGCU---------CGAGAAGCUACAGAGAUUCCGACUCCAAGGAUCUGG .(((.((((...................))))((((..(.(((((....))))).)...))))(((((...))---------)))..)))..((((...(((........))))))). ( -34.01) >DroAna_CAF1 24471 77 - 1 UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGC---------------------------------------CC-AACCGAAGGAACUC- ............(((.(((((......)).))).))).(.(((((....))))).)(((..---------------------------------------((-.......))..)))- ( -21.80) >consensus UGCUGACGUUAAUGACCGCCAAUAAAAUGCGUGCUCAAGUGGCAGGAAACUGCCUCGAGGAGCUACAGCGAUUCCAGC______________________CCGACUCCAAGGAUCUGG ((((.((((...................))))((((..(.(((((....))))).)...))))...))))....................................(((......))) (-19.55 = -20.61 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:43 2006