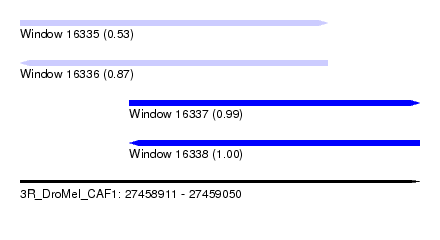
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,458,911 – 27,459,050 |
| Length | 139 |
| Max. P | 0.996109 |

| Location | 27,458,911 – 27,459,018 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27458911 107 + 27905053 AUUAUAUACCAGUCCUCACUGCUGCCAGACAUCCGUC--CAAUAGAGCUCGGCUCAGC-GGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGC----UA-AGAC ..........((..((....((((.(((((....)))--.....(((((((......)-))))))...(((((((.....))))))))).)))).....))..)----).-.... ( -32.00) >DroSec_CAF1 14792 100 + 1 ACUAUAUACCAGUCCUCACUGCUGCCAGACGUCCGUC--CAAUAGAGCUCGGCUCAGC-GGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCCUCUU------------UCG ....................((((.(((((....)))--.....(((((((......)-))))))...(((((((.....))))))))).))))......------------... ( -30.20) >DroSim_CAF1 14681 100 + 1 AUUAUAUACCAGUCCUCACUGCUGCCAGACGUCCGUC--CAAUAGAGCUCGGCUCAGC-GGGCUCAACGGAGUACAGUAAGUACUCUUGACAGCCUCGAU------------UCC ....................((((.(((((....)))--.....(((((((......)-))))))...(((((((.....))))))))).))))......------------... ( -27.50) >DroEre_CAF1 14749 107 + 1 AUUAUGUGCCAGUCCUCACUGCUGUCAGCCAUCCGUC--CAACAGAGGCGGGCUCAGC-GGGCUCAGCGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGC----UA-AGGA ........((((..((....((((((((((.((.(..--...).)))))(((((....-.)))))...(((((((.....)))))))))))))).....))..)----).-.)). ( -36.90) >DroYak_CAF1 14881 107 + 1 AUUAUAGCCCAGUCCUCACUGCUGCCAAACAUCCGUC--GAAUGGAGCCGGGCUCAGC-GGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGC----UG-AGGA ............((((((((((((((.....(((((.--..)))))....)))..)))-))((((...(((((((.....)))))))(((.....)))..))))----))-)))) ( -39.40) >DroPer_CAF1 17346 108 + 1 GUUAUUUCCAUGUACUCACUUAUGCCAGACUUCCUUGCUCAGCGGAGUC--GCUCAGUUGGGAGCUCUGAAGUACAGUGGACACUCCAG-----CACAAAGACCACGCUCAAUAC ......((((((((((..(....).((((.(((((.(((.((((....)--))).))).))))).)))).)))))).))))......((-----(...........)))...... ( -27.50) >consensus AUUAUAUACCAGUCCUCACUGCUGCCAGACAUCCGUC__CAAUAGAGCCCGGCUCAGC_GGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGC____U__AGAC .........((((....)))).......................((((...))))....(((((....(((((((.....)))))))....)))))................... (-16.58 = -17.45 + 0.87)



| Location | 27,458,911 – 27,459,018 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.24 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.07 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27458911 107 - 27905053 GUCU-UA----GCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCC-GCUGAGCCGAGCUCUAUUG--GACGGAUGUCUGGCAGCAGUGAGGACUGGUAUAUAAU ((((-(.----((((..((.((((....(((((((.....)))))))....)))))-)..))))...(((((.(..--(((....)))..).)).))).)))))........... ( -37.90) >DroSec_CAF1 14792 100 - 1 CGA------------AAGAGGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCC-GCUGAGCCGAGCUCUAUUG--GACGGACGUCUGGCAGCAGUGAGGACUGGUAUAUAGU (..------------..).(((((....(((((((.....)))))))....)))))-((((..((..(((((.(..--(((....)))..).)).)))..))..))))....... ( -38.30) >DroSim_CAF1 14681 100 - 1 GGA------------AUCGAGGCUGUCAAGAGUACUUACUGUACUCCGUUGAGCCC-GCUGAGCCGAGCUCUAUUG--GACGGACGUCUGGCAGCAGUGAGGACUGGUAUAUAAU (..------------.(((..(((((((.((((((.....))))))((((..(.((-(..((((...))))...))--).).))))..)))))))..)))...)........... ( -31.20) >DroEre_CAF1 14749 107 - 1 UCCU-UA----GCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGCUGAGCCC-GCUGAGCCCGCCUCUGUUG--GACGGAUGGCUGACAGCAGUGAGGACUGGCACAUAAU ...(-((----(.((((((..((((((((((((((.....)))))))(((.((...-.)).)))..(((((((...--..)))).))))))))))..))))))))))........ ( -44.50) >DroYak_CAF1 14881 107 - 1 UCCU-CA----GCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCC-GCUGAGCCCGGCUCCAUUC--GACGGAUGUUUGGCAGCAGUGAGGACUGGGCUAUAAU ..((-((----(.((((((..((((((((((((((.....)))))))((((((...-((((....))))....)))--))).......)))))))..)))))))))))....... ( -43.90) >DroPer_CAF1 17346 108 - 1 GUAUUGAGCGUGGUCUUUGUG-----CUGGAGUGUCCACUGUACUUCAGAGCUCCCAACUGAGC--GACUCCGCUGAGCAAGGAAGUCUGGCAUAAGUGAGUACAUGGAAAUAAC .........(((.((((((((-----(..((.(.(((.(((.....))).((((.......(((--(....))))))))..)))).))..))))))).)).)))........... ( -33.01) >consensus GCAU__A____GCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCC_GCUGAGCCGAGCUCUAUUG__GACGGAUGUCUGGCAGCAGUGAGGACUGGUAUAUAAU ....................((((....(((((((.....)))))))....)))).......(((..(((((.........))).))..)))..((((....))))......... (-22.20 = -22.07 + -0.13)

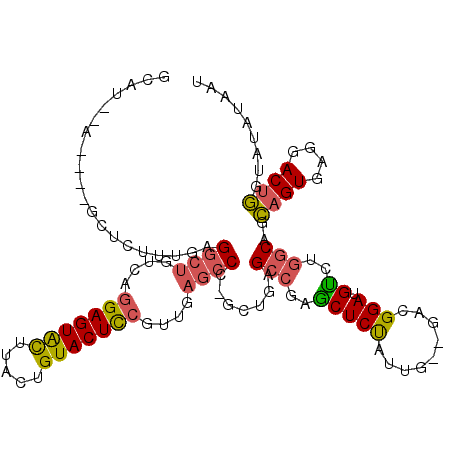
| Location | 27,458,949 – 27,459,050 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27458949 101 + 27905053 AAUAGAGCUCGGCUCAGCGGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGCUAAGAC-UUC-GAAGGAAUCAUUCAGUCGAACUGUUAAAC (((((..((..((((...(((((....(((((((.....)))))))....)))))....))))..))..-(((-((..((.....))..)))))))))).... ( -34.40) >DroSec_CAF1 14830 95 + 1 AAUAGAGCUCGGCUCAGCGGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCCUCUU-------UCGAGGC-UAAGGAAUCAUUCAGCCGAACUGUUAAAC (((((...((((((.((.(((((....(((((((.....)))))))....))))).)).-------..((..(-....)..))....)))))).))))).... ( -32.70) >DroSim_CAF1 14719 94 + 1 AAUAGAGCUCGGCUCAGCGGGCUCAACGGAGUACAGUAAGUACUCUUGACAGCCUCGAU-------UCC-UAC-GAAGGAAUCAUUCAGCCGAACUGUUAAAC (((((...((((((....(((((....(((((((.....)))))))....))))).(((-------(((-(..-..)))))))....)))))).))))).... ( -35.50) >DroEre_CAF1 14787 101 + 1 AACAGAGGCGGGCUCAGCGGGCUCAGCGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGCUAAGGA-UUC-GGAGGAAUCAUUCAGCCGAGCAGUUAAAC (((.((((((((((.....)))))...(((((((.....))))))).....)))))...(.(((...((-(((-....)))))....)))).....))).... ( -36.50) >DroYak_CAF1 14919 102 + 1 AAUGGAGCCGGGCUCAGCGGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGCUGAGGA-UUCAGGGGGAAUCAUUCAGCCGAACUGUUAAAC ....(((((.(......).)))))...(((((((.....)))))))((((((..((...(.((((((((-(((.....))))).))))))))).))))))... ( -38.00) >consensus AAUAGAGCUCGGCUCAGCGGGCUCAACGGAGUACAGUAAGUACUCCUGACAGCCUCAAAGAGCU_AGCA_UUC_GAAGGAAUCAUUCAGCCGAACUGUUAAAC (((((...((((((....(((((....(((((((.....)))))))....)))))......................(((....))))))))).))))).... (-24.56 = -24.80 + 0.24)


| Location | 27,458,949 – 27,459,050 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -26.21 |
| Energy contribution | -25.61 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27458949 101 - 27905053 GUUUAACAGUUCGACUGAAUGAUUCCUUC-GAA-GUCUUAGCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCCGCUGAGCCGAGCUCUAUU ........(((((((....((((..((((-(((-(........))))))))..))))(((((((.....)))))))))))))).....((((...)))).... ( -35.70) >DroSec_CAF1 14830 95 - 1 GUUUAACAGUUCGGCUGAAUGAUUCCUUA-GCCUCGA-------AAGAGGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCCGCUGAGCCGAGCUCUAUU .......(((((((((((.....))..((-((.((..-------..))(((((....(((((((.....)))))))....))))))))))))))))))..... ( -40.50) >DroSim_CAF1 14719 94 - 1 GUUUAACAGUUCGGCUGAAUGAUUCCUUC-GUA-GGA-------AUCGAGGCUGUCAAGAGUACUUACUGUACUCCGUUGAGCCCGCUGAGCCGAGCUCUAUU .......(((((((((.(..(((((((..-..)-)))-------)))(.((((.....((((((.....)))))).....)))))..).)))))))))..... ( -37.70) >DroEre_CAF1 14787 101 - 1 GUUUAACUGCUCGGCUGAAUGAUUCCUCC-GAA-UCCUUAGCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGCUGAGCCCGCUGAGCCCGCCUCUGUU ............((((((..(((((....-)))-)).))))))....(((((.....(((((((.....)))))))(((.((....)).)))..))))).... ( -35.90) >DroYak_CAF1 14919 102 - 1 GUUUAACAGUUCGGCUGAAUGAUUCCCCCUGAA-UCCUCAGCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCCGCUGAGCCCGGCUCCAUU ......(((...((((((..(((((.....)))-)).))))))..))).((((....(((((((.....)))))))....)))).((((....))))...... ( -34.50) >consensus GUUUAACAGUUCGGCUGAAUGAUUCCUUC_GAA_GCCU_AGCUCUUUGAGGCUGUCAGGAGUACUUACUGUACUCCGUUGAGCCCGCUGAGCCGAGCUCUAUU (((((.(.(((((((....((((.((((...................))))..))))(((((((.....))))))))))))))..).)))))........... (-26.21 = -25.61 + -0.60)

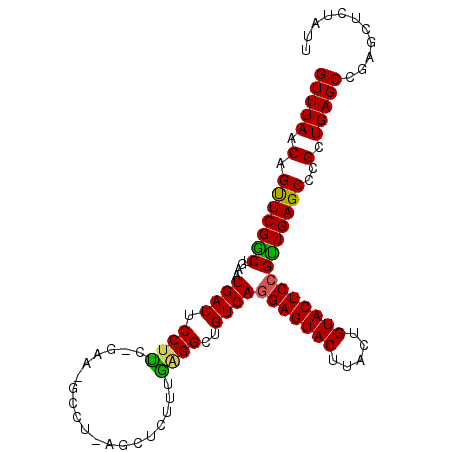
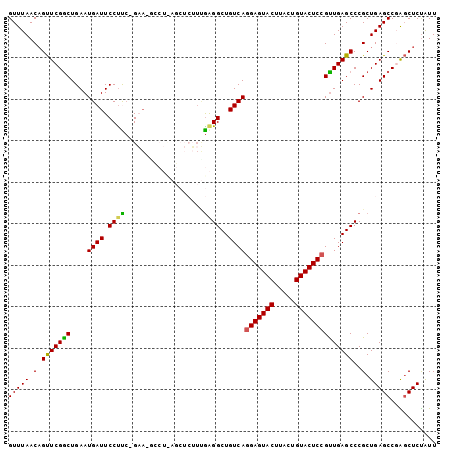
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:39 2006