
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,440,184 – 27,440,352 |
| Length | 168 |
| Max. P | 0.968003 |

| Location | 27,440,184 – 27,440,275 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27440184 91 - 27905053 UUGGUCGCCAGCGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCAAACACCUGCAUG-------AGAAAAUGUAUGU-UAAGCUAUCACAUUAA------ACUCU ...(((((((((((.....)).)))))))))......((((((.....))))))..(-------((..((((((((.-.....))).)))))..------.))). ( -30.10) >DroSec_CAF1 1565 92 - 1 UUGGUCGCCAACGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCGAACACCUGCAUG-------ACAAAUAUUAUGUUUAAGCAAUCACACUAC------ACUCU ...(((((((....(((((((....))).))))))))((((((.....))))))..)-------))........(((....)))..........------..... ( -25.20) >DroSim_CAF1 1639 92 - 1 UUGGUCGCCAACGAGUGCAUCUGCUGGUGGCACUGGCGCAGAUCAAACACCUGCAUG-------AGAAAAUUUAUGUUUAAGCAAUCACACUAC------ACUCU ((((((((...(.((((((((....))).))))).).)).))))))......(((((-------((....))))))).................------..... ( -18.90) >DroEre_CAF1 1561 98 - 1 UUGGUCGCCAACGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCAAACACCUGCAUG-------GCAAAUAUUGUGUUAAAGUAAUCGCAUUGAUUAGUAAUUCC ((((((((((.(.((((((((....))).))))).).((((((.....)))))).))-------))......((((..........))))..))))))....... ( -30.10) >DroYak_CAF1 1644 90 - 1 UUGGUCGCCAACGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCAAACACCUGUGUA---------UGAUAUUGGGUUAAAGUAAUUGCAUUGA------GAUCC ..((((.((((..((((((((....))).))))).((((((((.....)))))))).---------.....)))).((((.((....)).))))------)))). ( -26.10) >DroPer_CAF1 1565 96 - 1 UUGGUUGCCAGCGAAUGCAUCUGCUGGUGGCACUGACGCAAGUCAAACACCUGUUUAAAAAAUGGAUAAAAUUAUGA-----AAAUCAAGCUAA---GG-AAUCG ...((..((((((........))))))..))..((((....))))....((((((((.....))))).......((.-----....)).....)---))-..... ( -23.40) >consensus UUGGUCGCCAACGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCAAACACCUGCAUG_______ACAAAAAUUAUGUUUAAGCAAUCACACUAA______ACUCU (..((.(((.((.(((......))).)))))))..).((((((.....))))))................................................... (-18.17 = -18.08 + -0.08)

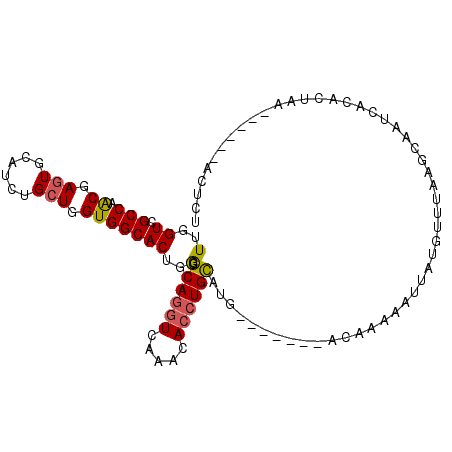
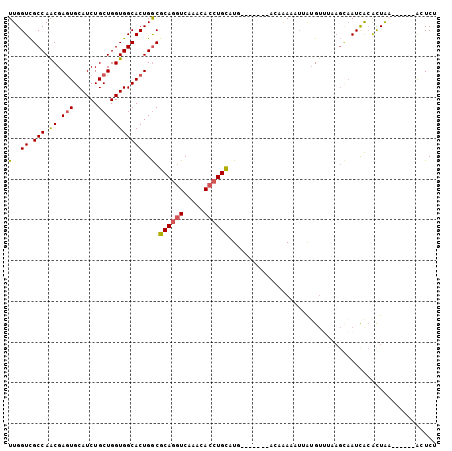
| Location | 27,440,205 – 27,440,315 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -30.91 |
| Energy contribution | -30.30 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27440205 110 + 27905053 ACAUACAUUUUCU--------CAUGCAGGUGUUUGACCUGCGCCAGUGCCACCAGCAGAUGCACUCGCUGGCGACCAAUGCGCAGGCAGCGGCCGCUGCUCAGGCGGCAGCCGUGGCC .............--------...((((((.....))))))(((..((((..((((((.....)).))))(((.......))).))))(((((.(((((....))))).)))))))). ( -47.60) >DroVir_CAF1 1549 110 + 1 CCGAAUCAU--------GUUUUCUGUAGGUCUUCGACUUGCGUCAAUGCCAUCAGCAGAUGCAUUCGCUGGCCACCAAUGCACAGGCCGCUGCUGCAGCGCAAGCGGCUGCCGUCGCG .(((....(--------((.(..((..((((..(((..((((((..(((.....))))))))).)))..))))..))..).)))(((.((((((((...)).)))))).))).))).. ( -38.10) >DroPse_CAF1 1581 115 + 1 UCAUAAUUUUCUCCAUUUUU-UA--CAGGUUUUUGACUUGCGUCAGUGCCACCAGCAGAUGCAUUCGCUGGCAACCAAUGCACAGGCAGCUGCCGCAGCACAGGCGGCCGCUGUGGCC .............((((...-..--..(((..(((((....))))).))).(((((.((.....))))))).....))))(((((.(.((((((........)))))).))))))... ( -35.90) >DroYak_CAF1 1666 108 + 1 ACCCAAUAUCA----------UACACAGGUGUUUGACCUGCGCCAGUGCCACCAGCAGAUGCACUCGUUGGCGACCAAUGCGCAGGCAGCGGCCGCAGCUCAGGCGGCAGCCGUGGCC ..((....(((----------.(((....))).)))(((((((...((.(.(((((((.....)).))))).)..))..)))))))..(((((.((.((....)).)).))))))).. ( -38.90) >DroMoj_CAF1 1541 105 + 1 UUUAAC--U--------GUU---AACAGGUGUUCGACUUGCGUCAAUGCCAUCAGCAGAUGCACUCGCUGGCCACCAAUGCACAGGCUGCUGCGGCAGCACAAGCGGCCGCCGUGGCA .....(--(--------((.---....((((..((.(.((((((..(((.....)))))))))...).))..)))).....))))((..(.(((((.((....)).))))).)..)). ( -39.80) >DroPer_CAF1 1584 117 + 1 UCAUAAUUUUAUCCAUUUUU-UAAACAGGUGUUUGACUUGCGUCAGUGCCACCAGCAGAUGCAUUCGCUGGCAACCAAUGCACAGGCAGCUGCCGCAGCACAGGCGGCCGCUGUGGCC ............((.....(-(((((....))))))(((((((..((....(((((.((.....)))))))..))..))))).))(((((.(((((.......))))).))))))).. ( -36.30) >consensus UCAUAAUUUU________UU_UAAACAGGUGUUUGACUUGCGUCAGUGCCACCAGCAGAUGCACUCGCUGGCAACCAAUGCACAGGCAGCUGCCGCAGCACAGGCGGCCGCCGUGGCC .........................(((((.....))))).((((((((..(((((.((.....)))))))........)))).(((.((((((........)))))).))).)))). (-30.91 = -30.30 + -0.61)



| Location | 27,440,205 – 27,440,315 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -45.63 |
| Consensus MFE | -33.55 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27440205 110 - 27905053 GGCCACGGCUGCCGCCUGAGCAGCGGCCGCUGCCUGCGCAUUGGUCGCCAGCGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCAAACACCUGCAUG--------AGAAAAUGUAUGU ((...(((((((.((....)).)))))))..((((((((....(((((((((((.....)).)))))))))....))))))))......))(((((.--------.....)))))... ( -53.20) >DroVir_CAF1 1549 110 - 1 CGCGACGGCAGCCGCUUGCGCUGCAGCAGCGGCCUGUGCAUUGGUGGCCAGCGAAUGCAUCUGCUGAUGGCAUUGACGCAAGUCGAAGACCUACAGAAAAC--------AUGAUUCGG ..((((((((((((((.((......))))))))..(((((((.((.....)).))))))).)))))..(((.(((((....))))).).))..........--------.....))). ( -41.80) >DroPse_CAF1 1581 115 - 1 GGCCACAGCGGCCGCCUGUGCUGCGGCAGCUGCCUGUGCAUUGGUUGCCAGCGAAUGCAUCUGCUGGUGGCACUGACGCAAGUCAAAAACCUG--UA-AAAAAUGGAGAAAAUUAUGA .(((((((((((.(((.((((.(((((....))).)))))).))).))).((....))....))).)))))..((((....))))....((..--..-......))............ ( -42.30) >DroYak_CAF1 1666 108 - 1 GGCCACGGCUGCCGCCUGAGCUGCGGCCGCUGCCUGCGCAUUGGUCGCCAACGAGUGCAUCUGCUGGUGGCACUGGCGCAGGUCAAACACCUGUGUA----------UGAUAUUGGGU (((..(((((((.((....)).)))))))..))).((.((...(((((((.(.(((......))).)))))....((((((((.....)))))))).----------.)))..)).)) ( -47.40) >DroMoj_CAF1 1541 105 - 1 UGCCACGGCGGCCGCUUGUGCUGCCGCAGCAGCCUGUGCAUUGGUGGCCAGCGAGUGCAUCUGCUGAUGGCAUUGACGCAAGUCGAACACCUGUU---AAC--------A--GUUAAA (((((((((((((....).)))))))((((((...(((((((.((.....)).))))))))))))).)))))(((((....)))))....(((..---..)--------)--)..... ( -46.60) >DroPer_CAF1 1584 117 - 1 GGCCACAGCGGCCGCCUGUGCUGCGGCAGCUGCCUGUGCAUUGGUUGCCAGCGAAUGCAUCUGCUGGUGGCACUGACGCAAGUCAAACACCUGUUUA-AAAAAUGGAUAAAAUUAUGA (((...(((.(((((.......))))).))))))(((.((((.((..((((((........))))))..))..((((....))))............-...)))).)))......... ( -42.50) >consensus GGCCACGGCGGCCGCCUGUGCUGCGGCAGCUGCCUGUGCAUUGGUCGCCAGCGAAUGCAUCUGCUGGUGGCACUGACGCAAGUCAAACACCUGUAUA_AA________AAAAUUAUGA .(((((.((.(((((.......))))).)).((..(((((((.((.....)).)))))))..))..)))))..((((....))))................................. (-33.55 = -33.88 + 0.34)


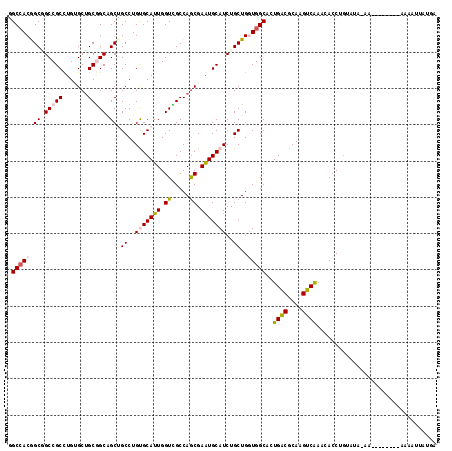
| Location | 27,440,235 – 27,440,352 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -50.23 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.12 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27440235 117 + 27905053 UGCGCCAGUGCCACCAGCAGAUGCACUCGCUGGCGACCAAUGCGCAGGCAGCGGCCGCUGCUCAGGCGGCAGCCGUGGCCGGAGUGGCCAAUCAGC---AAAUGGGCGGCGGCGGAAGGA ..((((((((......((....))...)))))))).((....(((.....)))((((((((((((((....))).(((((.....)))))......---...)))))))))))))..... ( -55.80) >DroVir_CAF1 1579 117 + 1 UGCGUCAAUGCCAUCAGCAGAUGCAUUCGCUGGCCACCAAUGCACAGGCCGCUGCUGCAGCGCAAGCGGCUGCCGUCGCGGGUGUCUCCAACCAAC---AAAUGGGCGGCGGCGGCAGAA ((((((..(((.....)))))))))..((((((((...........))))((((...))))...))))((((((((((((((....)))..(((..---...)))))))))))))).... ( -52.10) >DroGri_CAF1 1697 117 + 1 UGCGACAAUGCCAUCAGCAGAUGCACUCGCUGGCCACCAACGCACAAGCCGCUGCUGCAGCGCAAGCAGCUGCCGUCGCUGGUGUCUCCAAUCAAC---AAAUGGGCGGCGGUGGCAGAA .((((...(((.(((....)))))).))))..((((((.........((.((((((((...)).)))))).)).((((((.((.............---..)).)))))))))))).... ( -45.16) >DroWil_CAF1 1678 120 + 1 UGCGACAAUGUCACCAACAGAUGCACUCACUGGCCACCAAUGCCCAGGCUGCCGCAGCUGCACAAGCAGCCGCCGUUGCUGGCGUCUCCAAUCAACAACAGAUGGGCGGUGGCGGUAGAA ...(((...)))(((..(((.((....)))))((((((...(((((..(((..((.(((((....))))).)).((((.(((.....)))..))))..))).)))))))))))))).... ( -45.40) >DroMoj_CAF1 1566 117 + 1 UGCGUCAAUGCCAUCAGCAGAUGCACUCGCUGGCCACCAAUGCACAGGCUGCUGCGGCAGCACAAGCGGCCGCCGUGGCAGGCGUUUCCAAUCAAC---AAAUGGGCGGCGGUGGUAGAA ((((((..(((.....))))))))).((....((((((..(((...(.((((..((((.((.......)).))))..)))).)....(((......---...))))))..)))))).)). ( -47.80) >DroAna_CAF1 1588 117 + 1 UGCGCCAGUGCCACCAACAAAUGCAUUCUCUAGCCACGAAUGCCCAGGCAGCGGCGGCGGCUCAGGCGGCCGCAGUGGCCGGAGUGGCCAACCAGC---AGAUGGGUGGAGGCGGAAGAA ..((((....(((((......(((...(((..(((((...(((....)))...(((((.((....)).))))).)))))..)))(((....)))))---)....))))).))))...... ( -55.10) >consensus UGCGACAAUGCCACCAGCAGAUGCACUCGCUGGCCACCAAUGCACAGGCAGCUGCGGCAGCACAAGCGGCCGCCGUGGCCGGAGUCUCCAAUCAAC___AAAUGGGCGGCGGCGGAAGAA ((((....(((.....)))..)))).((((((.(((..........(((.((.((.((.((....)).)).)).)).)))((.....)).............))).))))))........ (-30.64 = -30.12 + -0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:25 2006