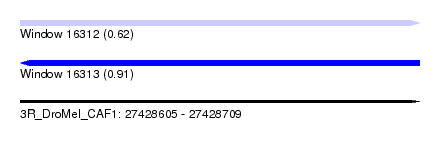
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,428,605 – 27,428,709 |
| Length | 104 |
| Max. P | 0.909940 |

| Location | 27,428,605 – 27,428,709 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -14.11 |
| Energy contribution | -16.50 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
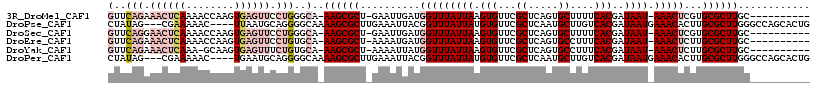
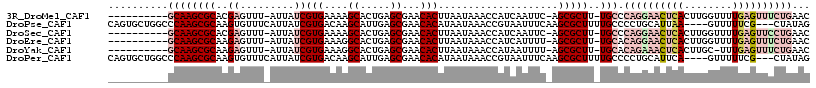
>3R_DroMel_CAF1 27428605 104 + 27905053 GUUCAGAAACUCAAAACCAAGUGAGUUCCUGGGCA-AAGCGCU-GAAUUGAUGGUUUAUUAAGUGUUCGCUCAGUGCUUUUCACGAUAAU-AAACUCGUGCGCUUGC---------- ((((((.((((((........)))))).)))))).-((((((.-......(((((((((((.(((...((.....))....)))..))))-)))).)))))))))..---------- ( -32.51) >DroPse_CAF1 4022 110 + 1 CUAUAG---CGAAAAAC----UUAAUGCAGGGGCAAAAGCGCUUGAAAUUACGGUUUAUUAUGUGUUCGCUCAAUGCUUGUCACGAUAAUGAAACACUUGCGCUUGGGCCAGCACUG ......---........----....(((...(((..((((((..(.........(((((((((((...((.....))....))).))))))))...)..))))))..))).)))... ( -26.20) >DroSec_CAF1 3581 104 + 1 GUUCAGGAACUCAAAACCAAGUGAGUUCCUGGGCA-AAGCGCU-GAAUUGAUGGUUUAUUAAGUGUUCGCUCAGUGCUUUUCACGAUAAU-AAACUCGUGCGCUUGC---------- (((((((((((((........))))))))))))).-((((((.-......(((((((((((.(((...((.....))....)))..))))-)))).)))))))))..---------- ( -39.31) >DroEre_CAF1 3886 104 + 1 GUUCAGAAACUCAAAACCAAGUGAGUUCCUGUGCA-AAGCGCU-AAAAUGAUGGUUUAUUAAGUGUUCGCUCAGUGCCUUUCACGAUAAU-AAACUCUUGCGCUUGC---------- ((.(((.((((((........)))))).))).)).-((((((.-.....((.(((((((((.(((...((.....))....)))..))))-))))).))))))))..---------- ( -29.00) >DroYak_CAF1 6543 103 + 1 GUUCAGAAACUCAAA-GCAAGUGAGUUUCUGUGCA-AAGCGCU-AAAAUUAUGGUUUAUUAAGUGUUCGCUCAGUGCCUUUCACGAUAAU-AAACUCUUGCGCUUGC---------- ((.((((((((((..-.....)))))))))).)).-((((((.-........(((((((((.(((...((.....))....)))..))))-)))))...))))))..---------- ( -32.70) >DroPer_CAF1 4036 110 + 1 CUAUAG---CGAAAAAC----UGAAUGCAGGGGCAAAAGCGCUUGAAAUUACGGUUUAUUAUGUGUUCGCUCAAUGCUUGUCACGAUAAUGAAACACUUGCGCUUGGGCCAGCACUG .....(---(......(----((....))).(((..((((((..(.........(((((((((((...((.....))....))).))))))))...)..))))))..))).)).... ( -28.00) >consensus GUUCAGAAACUCAAAACCAAGUGAGUUCCUGGGCA_AAGCGCU_GAAAUGAUGGUUUAUUAAGUGUUCGCUCAGUGCUUUUCACGAUAAU_AAACUCUUGCGCUUGC__________ (..(((.((((((........)))))).)))..)..((((((..........(((((((((.(((...((.....))....)))..)))).)))))...))))))............ (-14.11 = -16.50 + 2.39)
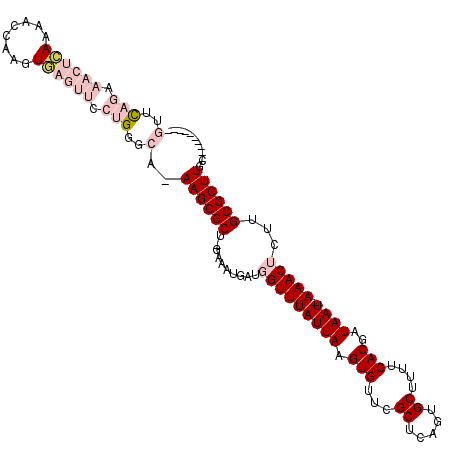
| Location | 27,428,605 – 27,428,709 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -15.66 |
| Energy contribution | -18.22 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
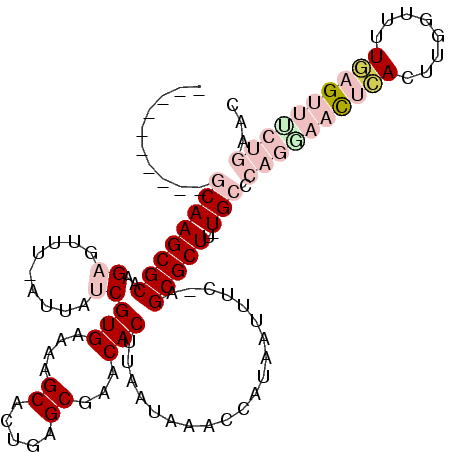
>3R_DroMel_CAF1 27428605 104 - 27905053 ----------GCAAGCGCACGAGUUU-AUUAUCGUGAAAAGCACUGAGCGAACACUUAAUAAACCAUCAAUUC-AGCGCUU-UGCCCAGGAACUCACUUGGUUUUGAGUUUCUGAAC ----------((((((((..((((((-((((..(((....((.....))...))).)))))))).......))-.)))).)-))).((((((((((........))))))))))... ( -30.61) >DroPse_CAF1 4022 110 - 1 CAGUGCUGGCCCAAGCGCAAGUGUUUCAUUAUCGUGACAAGCAUUGAGCGAACACAUAAUAAACCGUAAUUUCAAGCGCUUUUGCCCCUGCAUUAA----GUUUUUCG---CUAUAG .(((((.(((..((((((..(.((((.(((((.(((....((.....))...)))))))))))))(......)..))))))..)))...)))))..----........---...... ( -25.80) >DroSec_CAF1 3581 104 - 1 ----------GCAAGCGCACGAGUUU-AUUAUCGUGAAAAGCACUGAGCGAACACUUAAUAAACCAUCAAUUC-AGCGCUU-UGCCCAGGAACUCACUUGGUUUUGAGUUCCUGAAC ----------((((((((..((((((-((((..(((....((.....))...))).)))))))).......))-.)))).)-))).((((((((((........))))))))))... ( -33.51) >DroEre_CAF1 3886 104 - 1 ----------GCAAGCGCAAGAGUUU-AUUAUCGUGAAAGGCACUGAGCGAACACUUAAUAAACCAUCAUUUU-AGCGCUU-UGCACAGGAACUCACUUGGUUUUGAGUUUCUGAAC ----------((((((((((((((((-((((..(((....((.....))...))).)))))))).....))))-.)))).)-))).((((((((((........))))))))))... ( -32.10) >DroYak_CAF1 6543 103 - 1 ----------GCAAGCGCAAGAGUUU-AUUAUCGUGAAAGGCACUGAGCGAACACUUAAUAAACCAUAAUUUU-AGCGCUU-UGCACAGAAACUCACUUGC-UUUGAGUUUCUGAAC ----------((((((((((((((((-((((..(((....((.....))...))).)))))))).....))))-.)))).)-))).((((((((((.....-..))))))))))... ( -32.30) >DroPer_CAF1 4036 110 - 1 CAGUGCUGGCCCAAGCGCAAGUGUUUCAUUAUCGUGACAAGCAUUGAGCGAACACAUAAUAAACCGUAAUUUCAAGCGCUUUUGCCCCUGCAUUCA----GUUUUUCG---CUAUAG .(((((.(((..((((((..(.((((.(((((.(((....((.....))...)))))))))))))(......)..))))))..)))...)))))..----........---...... ( -25.20) >consensus __________GCAAGCGCAAGAGUUU_AUUAUCGUGAAAAGCACUGAGCGAACACUUAAUAAACCAUAAUUUC_AGCGCUU_UGCCCAGGAACUCACUUGGUUUUGAGUUUCUGAAC ..........((((((((..((.........))(((....((.....))...)))....................)))))..))).((((((((((........))))))))))... (-15.66 = -18.22 + 2.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:17 2006