
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,428,007 – 27,428,135 |
| Length | 128 |
| Max. P | 0.935817 |

| Location | 27,428,007 – 27,428,109 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
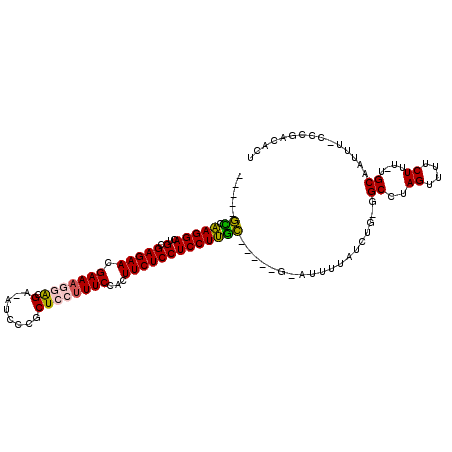
>3R_DroMel_CAF1 27428007 102 - 27905053 -----GCCAAGGACUGCGAGAACGAAAGGACGA-AUUCCGCUCCCUUCCACUUCUCCUCCUUGC-----G-AUUUCAUCUG-GGCCUAGUUUUCUUUUUGCACUUU-CCCGACACU -----((.(((((..(.(((((.(((.(((((.-....)).))).)))...)))))))))))))-----.-........((-((...(((...........)))..-))))..... ( -21.20) >DroPse_CAF1 3320 106 - 1 AG---AGAGAGGACUGCGAGAACGAAAGCGAGACAUCCUGCUCAUUUCCACUUCUCCUCCUCCUCAUACGCAUAUUGUGUGUGGCCUAGUUUUCUUU-UGCAA------CGGCACU .(---(((((((((((((((((.((((..(((.(.....)))).))))...)))))........(((((((.....)))))))))..))))))))))-)((..------..))... ( -29.80) >DroEre_CAF1 3280 102 - 1 -----GCCAAGGACUGCGAGAACGAAAGGACGA-AUUCCGCUCCUUUCCACUUCUCCUCCUUGC-----G-AUUUUAUCUG-GGCCUAGUUUUCUUUUUGCACUUU-CCCGACACU -----((.(((((..(.(((((.(((((((((.-....)).)))))))...)))))))))))))-----.-........((-((...(((...........)))..-))))..... ( -25.30) >DroYak_CAF1 5938 102 - 1 -----GCCAAGGACUGCGAGAACGAAAGGACGA-AUACCGCUCCUUUCCACUUCUCCUCCUUGC-----G-AUUUUCUCUG-GGCCUAGUUUUCUUUUUGCACUUU-CCCGACACU -----((.(((((..(.(((((.(((((((((.-....)).)))))))...)))))))))))))-----.-........((-((...(((...........)))..-))))..... ( -25.30) >DroAna_CAF1 2891 104 - 1 UGGAAGUCAAGGACUGCGAGAACGAAAGGACGA-GCCCCUCUCCUUUCCACUUCUCCUCCUUGCCC---G-AUU-----CA-GGCCUAGUUUUCUUU-UGCAAUCUCCUCGACACU .....(((.((((.((((((((.(((((((.((-(...))))))))))...)))))...((.(((.---.-...-----..-)))..))........-.)))...)))).)))... ( -29.00) >DroPer_CAF1 3334 106 - 1 AG---AGAGAGGACUGCGAGAACGAAAGCGAGACAUCCUGCUCAUUUCCACUUCUCCUCCUCCUCAUACGCAUAUUGUGUGUGGCCUAGUUUUCUUU-UGCAA------CGGCACU .(---(((((((((((((((((.((((..(((.(.....)))).))))...)))))........(((((((.....)))))))))..))))))))))-)((..------..))... ( -29.80) >consensus _____GCCAAGGACUGCGAGAACGAAAGGACGA_AUCCCGCUCCUUUCCACUUCUCCUCCUUGC_____G_AUUUUAUCUG_GGCCUAGUUUUCUUU_UGCAAUUU_CCCGACACU .....((.(((((..(.(((((.(((((((.(........))))))))...)))))))))))))...................((..((....))....))............... (-15.10 = -14.30 + -0.80)

| Location | 27,428,045 – 27,428,135 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.94 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -15.00 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
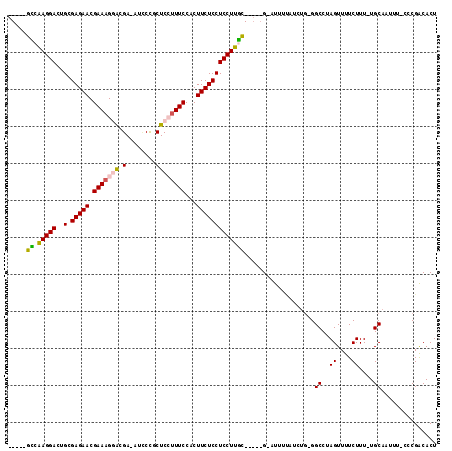
>3R_DroMel_CAF1 27428045 90 - 27905053 GCUGAAAAUAAAAG-CAAACAAAAUUU-----GCCAAGGACUGCGAGAACGAAAGGACGA-AUUCCGCUCCCUUCCACUUCUCCUCCUUGC-----G-AUUUC (((.........))-).........((-----((.(((((..(.(((((.(((.(((((.-....)).))).)))...)))))))))))))-----)-).... ( -20.80) >DroPse_CAF1 3353 100 - 1 GCUGAAAAUAAAAGUCAAACAAGCGAGAG---AGAGAGGACUGCGAGAACGAAAGCGAGACAUCCUGCUCAUUUCCACUUCUCCUCCUCCUCAUACGCAUAUU ......................(((...(---((.(((((..(.(((((.((((..(((.(.....)))).))))...))))))))))))))...)))..... ( -23.60) >DroEre_CAF1 3318 90 - 1 GCUGAAAAUAAAAG-CAAACAAAAUUU-----GCCAAGGACUGCGAGAACGAAAGGACGA-AUUCCGCUCCUUUCCACUUCUCCUCCUUGC-----G-AUUUU (((.........))-).....(((.((-----((.(((((..(.(((((.(((((((((.-....)).)))))))...)))))))))))))-----)-).))) ( -25.00) >DroYak_CAF1 5976 90 - 1 GCUGAAAAUAAAAG-CAAACAAAAUUU-----GCCAAGGACUGCGAGAACGAAAGGACGA-AUACCGCUCCUUUCCACUUCUCCUCCUUGC-----G-AUUUU (((.........))-).....(((.((-----((.(((((..(.(((((.(((((((((.-....)).)))))))...)))))))))))))-----)-).))) ( -25.00) >DroAna_CAF1 2926 95 - 1 GCUGAAAAUAGAAU-CAAACAAUCUUUUGGAAGUCAAGGACUGCGAGAACGAAAGGACGA-GCCCCUCUCCUUUCCACUUCUCCUCCUUGCCC---G-AUU-- ..............-...........((((..(.((((((..(.(((((.(((((((.((-(...))))))))))...)))))))))))))))---)-)..-- ( -23.60) >DroPer_CAF1 3367 100 - 1 GCUGAAAAUAAAAGUCAAACAAGCGAGAG---AGAGAGGACUGCGAGAACGAAAGCGAGACAUCCUGCUCAUUUCCACUUCUCCUCCUCCUCAUACGCAUAUU ......................(((...(---((.(((((..(.(((((.((((..(((.(.....)))).))))...))))))))))))))...)))..... ( -23.60) >consensus GCUGAAAAUAAAAG_CAAACAAAAUUU_____GCCAAGGACUGCGAGAACGAAAGGACGA_AUCCCGCUCCUUUCCACUUCUCCUCCUUGC_____G_AUUUU ................................((.(((((..(.(((((.(((((((.(........))))))))...)))))))))))))............ (-15.00 = -14.20 + -0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:15 2006