
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 27,422,605 – 27,422,753 |
| Length | 148 |
| Max. P | 0.992807 |

| Location | 27,422,605 – 27,422,713 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -18.63 |
| Consensus MFE | -11.00 |
| Energy contribution | -12.00 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27422605 108 + 27905053 CCUAA-AGUGUAAACCUCAACCGAAUAUAAGCUCAAAACUUGACGAUGGGCAAUUGCCCACCAUCAUUGUUAUCAUGAAUAAAAGUAGUCGUCAAAUAAAAACUAAAUA----------- .....-.................................((((((((((((....))))....((((.......)))).........))))))))..............----------- ( -18.40) >DroSim_CAF1 257 108 + 1 CCUAA-AAUGUAAACCUCAACCGAAUAUAAGCUCAAAACUUGACGAUGGGCGAUUGCCCACCAUCAUUGUUAGCAUCAAUAAAAGUAGUCGUCAAAUAAAAACUAAAUA----------- .....-.................................((((((((((((....))))......((((.......)))).......))))))))..............----------- ( -17.50) >DroYak_CAF1 361 118 + 1 CCUAAAAAUGUAAACCUCAACCGAAUAUAAGCUCAAAACUUGCCGAUGGUGGAUUAACCAUC--CAUCAUCAGCAUCAAUAAAACUAGUCAUCAAAUAAAAACUAAUUAUGCAUACACUU .......(((((............................(((.((((((((((.....)))--))))))).)))..........((((............))))....)))))...... ( -20.00) >consensus CCUAA_AAUGUAAACCUCAACCGAAUAUAAGCUCAAAACUUGACGAUGGGCGAUUGCCCACCAUCAUUGUUAGCAUCAAUAAAAGUAGUCGUCAAAUAAAAACUAAAUA___________ .......................................((((((((((((....)))).......(((((......))))).....))))))))......................... (-11.00 = -12.00 + 1.00)



| Location | 27,422,605 – 27,422,713 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.67 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27422605 108 - 27905053 -----------UAUUUAGUUUUUAUUUGACGACUACUUUUAUUCAUGAUAACAAUGAUGGUGGGCAAUUGCCCAUCGUCAAGUUUUGAGCUUAUAUUCGGUUGAGGUUUACACU-UUAGG -----------.....(((((..(((((((............((((.......)))).(((((((....))))))))))))))...))))).........(((((((....)))-)))). ( -28.10) >DroSim_CAF1 257 108 - 1 -----------UAUUUAGUUUUUAUUUGACGACUACUUUUAUUGAUGCUAACAAUGAUGGUGGGCAAUCGCCCAUCGUCAAGUUUUGAGCUUAUAUUCGGUUGAGGUUUACAUU-UUAGG -----------.....(((((..(((((((((..(((.((((((.......)))))).)))((((....)))).)))))))))...))))).........(((((((....)))-)))). ( -26.70) >DroYak_CAF1 361 118 - 1 AAGUGUAUGCAUAAUUAGUUUUUAUUUGAUGACUAGUUUUAUUGAUGCUGAUGAUG--GAUGGUUAAUCCACCAUCGGCAAGUUUUGAGCUUAUAUUCGGUUGAGGUUUACAUUUUUAGG (((((((.((..((((((((.((....)).))))))))..(((((.(((((((.((--(((.....))))).)))))))((((.....))))....)))))....)).)))))))..... ( -30.10) >consensus ___________UAUUUAGUUUUUAUUUGACGACUACUUUUAUUGAUGCUAACAAUGAUGGUGGGCAAUCGCCCAUCGUCAAGUUUUGAGCUUAUAUUCGGUUGAGGUUUACAUU_UUAGG ...............(((((.((....)).)))))...................(((((((((((....))))))))))).((...((.((((........)))).)).))......... (-20.33 = -20.67 + 0.34)



| Location | 27,422,644 – 27,422,753 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27422644 109 + 27905053 UGACGAUGGGCAAUUGCCCACCAUCAUUGUUAUCAUGAAUAAAAGUAGUCGUCAAAUAAAAACUAAAUA-----------UACAAUAAUUGAGAUUUUCAUUCAUGUUUUUGAUGUUGAU (((((((((((....))))....((((.......)))).........)))))))...............-----------..(((((.(..(((.............)))..)))))).. ( -20.82) >DroSim_CAF1 296 109 + 1 UGACGAUGGGCGAUUGCCCACCAUCAUUGUUAGCAUCAAUAAAAGUAGUCGUCAAAUAAAAACUAAAUA-----------UACAAUAAUUGAGAUUUUCAUUCAUGUUUUUGAUGUUGAU (((.(.(((((....))))).).)))..(((((((((((......((((..(......)..))))....-----------............((.......))......))))))))))) ( -20.10) >DroYak_CAF1 401 118 + 1 UGCCGAUGGUGGAUUAACCAUC--CAUCAUCAGCAUCAAUAAAACUAGUCAUCAAAUAAAAACUAAUUAUGCAUACACUUUACAAUAAUUGAGAUUUUUCUUCACGUUUUUGAUGUUGAU ....((((((((.....))).)--))))(((((((((((..((((...................((((((..............))))))((((....))))...))))))))))))))) ( -25.74) >consensus UGACGAUGGGCGAUUGCCCACCAUCAUUGUUAGCAUCAAUAAAAGUAGUCGUCAAAUAAAAACUAAAUA___________UACAAUAAUUGAGAUUUUCAUUCAUGUUUUUGAUGUUGAU (((.(.(((((....))))).).)))..(((((((((((......((((............))))...........................((.......))......))))))))))) (-15.12 = -15.57 + 0.45)

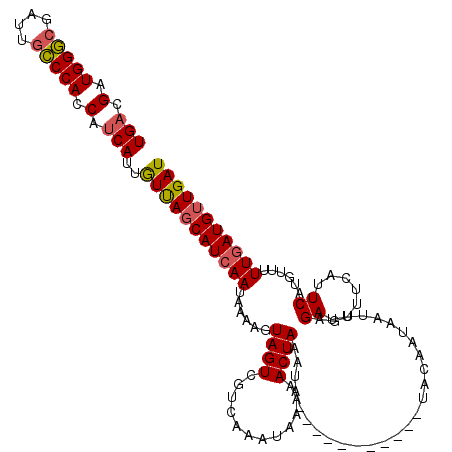

| Location | 27,422,644 – 27,422,753 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -19.85 |
| Energy contribution | -20.97 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 27422644 109 - 27905053 AUCAACAUCAAAAACAUGAAUGAAAAUCUCAAUUAUUGUA-----------UAUUUAGUUUUUAUUUGACGACUACUUUUAUUCAUGAUAACAAUGAUGGUGGGCAAUUGCCCAUCGUCA ..............(((((((((((.((((((..((((..-----------....))))......)))).))....))))))))))).......(((((((((((....))))))))))) ( -29.50) >DroSim_CAF1 296 109 - 1 AUCAACAUCAAAAACAUGAAUGAAAAUCUCAAUUAUUGUA-----------UAUUUAGUUUUUAUUUGACGACUACUUUUAUUGAUGCUAACAAUGAUGGUGGGCAAUCGCCCAUCGUCA .....((((((..((((((.(((.....))).))).))).-----------....(((((.((....)).)))))......)))))).......(((((((((((....))))))))))) ( -27.20) >DroYak_CAF1 401 118 - 1 AUCAACAUCAAAAACGUGAAGAAAAAUCUCAAUUAUUGUAAAGUGUAUGCAUAAUUAGUUUUUAUUUGAUGACUAGUUUUAUUGAUGCUGAUGAUG--GAUGGUUAAUCCACCAUCGGCA .....(((((((..(.....)(((((.((.((((((.(((.......)))))))))))))))).)))))))..............((((((((.((--(((.....))))).)))))))) ( -27.00) >consensus AUCAACAUCAAAAACAUGAAUGAAAAUCUCAAUUAUUGUA___________UAUUUAGUUUUUAUUUGACGACUACUUUUAUUGAUGCUAACAAUGAUGGUGGGCAAUCGCCCAUCGUCA .....((((((..((((((.(((.....))).))).)))................(((((.((....)).)))))......)))))).......(((((((((((....))))))))))) (-19.85 = -20.97 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:52:08 2006